Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA LINC01783 Promotes the Progression of Cervical Cancer by Sponging miR-199b-5p to Mediate GBP1 Expression
Authors Chen W, Xiong L, Yang L, Yang L, Li L, Huang L, Liang X, Xue J, Tan B
Received 7 September 2019
Accepted for publication 1 November 2019
Published 16 January 2020 Volume 2020:12 Pages 363—373
DOI https://doi.org/10.2147/CMAR.S230171
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
This paper has been retracted.
Wei-jun Chen, 1,* Liang Xiong, 2,* Lin Yang, 3 Li-juan Yang, 3 Lin Li, 3 Li Huang, 3 Xiao-qing Liang, 4 Jie Xue, 3 Bu-zhen Tan 5
1Department of Gynaecology and Obstetrics, The Second Affiliated Hospital of Nanchang University, Reproductive Hospital Affiliated to Jiangxi University of Traditional Chinese Medicine, Nanchang, Jiangxi, People’s Republic of China; 2Department of Urinary Surgery, Armed Police Jiangxi General Team Hospital, Nanchang, Jiangxi, People’s Republic of China; 3Department of Reproductive Medicine, Reproductive Hospital Affiliated to Jiangxi University of Traditional Chinese Medicine, Nanchang, Jiangxi, People’s Republic of China; 4Department of Gynaecology and Obstetrics, The Fourth Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, People’s Republic of China; 5Department of Gynaecology and Obstetrics, The Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bu-zhen Tan Email [email protected]
Background: Long non-coding RNA showed potential regulating effects in oncogenesis. Highly expressed LncRNA LINC01783 is observed in cervical cancer. However, the specific pathogenesis of cervical cancer is still unclear.
Methods: Differential lncRNAs in cervical cancer were identified based on TCGA dataset. Subsequently, qRT-PCR was utilized for testing the LINC01783 expression in cervical cancer cell lines and normal human cervical epithelial cell line HcerEpic. CCK-8, EdU, Wound healing assay, Transwell assay and flow cytometry were used for detecting proliferative and migratory potential, cell cycle and apoptosis of cervical cancer cells, respectively. To identify the potential target of LINC01783, bioinformatics assay and dual-luciferase reporter gene assay were performed. Moreover, to clarify their interactions and roles in regulating the progression of cervical cancer, Western blot assay and RIP assay were carried out.
Results: Our results revealed LINC01783 is overexpressed in cervical cancer cells. Overexpressed LINC01783 considerably accelerated the cell proliferation, migration and invasion of cervical cancer cells while restrained cell apoptosis of them. Moreover, LINC01783 positively regulated the GBP1 expression via competitively binding to miR-199b-5p.
Conclusion: LINC01783 is involved in the progression of cervical cancer through competitively binding to miR-199b-5p to mediate GBP1 expression.
Keywords: cervical cancer, LncRNA, ceRNA, proliferation, migration, invasion
Introduction
Cervical cancer is still the third most common form of cancers in developing countries1 with a five-year survival rate of 17%,2 albeit with extensive screening schemes. Cervical cancer may occur if an individual persistently infects with a high-risk strain of HPV, mainly including HPV 16 and HPV 18.3 It recurs in one-third of female patients treated, almost ineluctably resulting in fatal outcome.4 Long non-coding RNAs (lncRNAs) were reported to be related to the progression of cervical cancer. For instance, the study of Yan et al found that the proliferative and invasive potentials of cervical cancer cells were restrained by lncRNA UCA1 downregulation via miR-206 expression.5 In addition, Wen et al suggested that long noncoding RNA GAS5 regulated the expression of cisplatin resistance in cervical cancer as a tumor suppressor via microRNA 21.6 This study was designed to elaborate the epigenetic mechanism of the occurrence, progression, metastasis and invasion of cervical cancer. Our results are valuable for the improvement of the diagnosis and treatment of cervical cancer.
Long non-coding RNAs (lncRNAs) are non-coding RNAs that contain >200 nucleotides and may regulate the gene expression.7,8 As a result of complex biological effects, lncRNAs have gained much attention over the years. Certain lncRNAs have been reported crucial in the proliferation, apoptosis, infiltration and invasion of many tumor cells.6,9,10 LncRNA LINC01783 (Gene ID: 100132147) locates in the 1p36.13 region of human genome. The role of LINC01783 has not been reported so far.
In this study, the TCGA data were screened to identify differentially expressed lncRNAs in cervical cancer tissues and normal tissues, and cervical cancer cell lines were collected for quantitative real-time PCR (qRT-PCR). LINC01783 was selected as the object of research. Our results confirmed highly expressed LINC01783 in cervical cancer cell lines. LINC01783 overexpression accelerated the proliferation, migration, invasion and cycle of HeLa and C-33A cells and suppressed the apoptosis of HeLa and C-33A cells. In summary, it is proven that LINC01783 is involved in the cervical cancer progression through competitively binding to miR-199b-5p to mediate GBP1 (guanylate binding protein 1) expression.
Materials and Methods
Cell Culture and Transfection
Cervical cancer cell lines (SW756, C-33A, SiHa, HeLa and CaSki) and normal human cervical epithelial cell line (HcerEpic) were obtained from ATCC (Manassas VA, USA). Cell culture was conducted in DMEM mixed with 10% FBS (Beyotime, Nantong, China), 100 μg/mL streptomycin and 100 IU/mL penicillin (Invitrogen, USA), followed by preservation in 5% CO2 at 37°C. GenePharma (Shanghai, China) constructed LINC01783 overexpression plasmid, LINC01783 siRNA, miR-199b-5p mimics and miR-199b-5p inhibitor. Cells were transfected with Lipofectamine 2000 (Invitrogen, CA, USA).
RNA Extraction and qRT-PCR
Reverse Transcription Kit (Takara, Tokyo, Japan) was utilized for reversely transcribing RNAs into cDNAs while 2−ΔΔCt method was used for RNA quantification via normalizing to GAPDH. GBP1 primer sequences were shown below: F: 5ʹ- AGGAGTTCCTTCAAAGATGTGGA-3ʹ, R: 5ʹ-GCAACTGGACCCTGTCGTT-3ʹ. LINC01783 primer sequences were shown below: F: 5ʹ-CAAGGACAGCAGGTGGAGTA-3ʹ, R: 5ʹ-CTTACAGTGGACTCGGGGTT-3ʹ. All these experiments were separately conducted for three times.
Cell Proliferation Assay
Cells were subject to culture in 96-well plates and 1 h of incubation using CCK-8 reagent (Beyotime, Nantong, China). Next, TECAN infinite M200 Multimode microplate reader (Tecan, Mechelen, Belgium) was applied for absorbance recording at 450nm.
In regard to EDU assay, EdU reagent was used for 2 h of cell culture. Before EdU staining as recommended by the manufacturer, 15 min of cell fixation was conducted in 4% paraformaldehyde.
Wound Healing Assay
After seeding transfected cells into 6-well plates, they were grown until 100% confluence in DMEM medium. Then, a pipette tip was used for creating wounds on cell monolayer layers. Cell washing was performed using PBS, followed by 24 h incubation in free-serum medium. At 0 h and 24 h, AxioVision v4.7 software (Carl Zeiss Meditec, Dublin, CA, USA) and microscope were used for the measurement and imaging of wound closure, respectively.
Transwell Invasion Assay
Cell invasion was detected by Transwell insert chamber (Corning, NY, USA) with Matrigel coating (BD Biosciences, Franklin Lakes, NJ, USA). The upper chamber was seeded with transfected cells at 2.0×104 cells/well in serum-free medium, while the lower chamber included DMEM medium with 10% FBS as chemoattractant. After incubation for 48 h at 37°C, cotton swabs were used for wiping off the non-invaded cells on the top of Transwell. The invaded cells on the lower portion were subject to 20 min fixation in 4% paraformaldehyde and 15 min staining using 1% crystal violet (Sigma). In addition, under a light microscope (Olympus), the invaded cells in five random fields per filter were imaged and counted.
Cell Apoptosis and Cell Cycle Assay
Propidium iodide (PI) and Annexin V-FITC were used for cell dyeing, while flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) was used for determining the cell apoptosis.
PI and flow cytometry were used for cell staining and determination, respectively, while flow cytometer (FACScan; BD Biosciences, USA) installed with Cell Quest software (BD Biosciences) was performed to analyze the cell cycle.
Subcellular Distribution
PARIS Kit (Life Technologies, USA) was applied for extracting RNAs in cytoplasm and nucleus, while qRT-RCR was used for the quantification of total RNA in each fraction. Nucleus and cytoplasm internal references were U6 and GAPDH, respectively.
Dual-Luciferase Reporter Gene Assay
Mutant-type plasmids of LINC01783 MUT and GBP1 MUT and wild-type plasmids of LINC01783 WT and GBP1 WT were constructed. After seeding HeLa and C-33A cells into 24-well plates, Lipofectamine 2000 was used for co-transfection with 50 nM miR-199b-5p mimics or negative control and mutant- or wild-type plasmid. Every 80 ng of plasmid was added with 5 ng of pRL-SV40. On a microplate reader, luciferase intensity was detected with dual-luciferase reporter assay kit (Promega, Madison, WI, USA).
RNA Immunoprecipitation (RIP) Assay
RIP assay was performed using Magna Nuclear RIP™ (Native) Nuclear RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Then, cell lysis was conducted in complete RIPA buffer with RNase inhibitor and protease inhibitor cocktail. Cell extracts were incubated by RIP buffer with magnetic beads post conjugation with IgG control or human anti-Ago2 antibody (Millipore). Immunoprecipitated RNAs were established by protein digestion. Ultimately, qRT-PCR was performed to quantify the purified RNAs. Anti-LINC01783 used in RIP assay was commercially available from Abcam (Cambridge, MA, USA).
Western Blot
BCA method was used for the extraction and quantification of protein samples, followed by separation using SDS-PAGE gel electrophoresis and blocking using 5% skim milk. Subsequently, membrane incubation was performed using rabbit anti-human IgG antibodies against GBP1 and GAPDH as primary antibodies and their secondary antibodies. Chemiluminescence was used for the development of band exposure.
Bioinformatics Analysis
For The cancer genome atlas (TCGA) data, the TCGA (https://portal.gdc.cancer.gov/) was used for identifying gene expression difference. P < 0.05 and the log2 fold change (log2FC) >1 or <−1 was considered statistically significant.
Statistical Processing
All statistical analyses in the present study were performed with GraphPad Prism 6.0 and SPSS 20.0 software. Quantitative data were indicated as mean ± SD. T-test was made for analyzing measurement data, whereas nonparametric test was used for data not in normal distribution. P<0.05 indicated statistically significant difference.
Results
LINC01783 Expression in Cervical Cancer
To identify lncRNAs associated with cervical cancer, we analyzed the lncRNA expression on TCGA dataset. In total, we found 1979 dysregulated lncRNAs including 927 upregulated lncRNAs and 1052 downregulated lncRNAs in cervical cancer tissues (Supplementary Figure 1A, B). As shown in Figure 1A, LINC01783 has high levels in cervical cancer tissues according to TCGA. Overall survival of cervical cancer patients stratified by LINC01783 expression based on TCGA dataset showed that the overall survival rate of cervical cancer patients with high expression of LINC01783 was lower than it of the low-expression group (Figure 1B). In Figure 1C, qRT-PCR was utilized for detecting LINC01783 expression in cervical cancer cell lines (SW756, C-33A, SiHa, CaSki and HeLa) and normal human cervical epithelial cell (HcerEpic).
LINC01783 Functions in Cervical Cancer Cell Lines
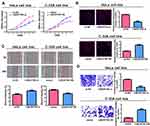
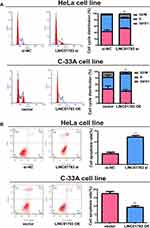
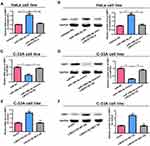
Among the selected cervical cancer cell lines for subsequent experiments, the greatest LINC01783 expression was observed in HeLa cells whereas the lowest expression was observed in C-33A cells. Furthermore, qRT-PCR (Supplementary Figure 2A, B) was used for the verification of transfection efficacy of LINC01783 siRNA and LINC01783 overexpression vector in cervical cancer cells. Meanwhile, the proliferation abilities of cervical cancer cells were considerably decreased by downregulated LINC01783 as indicated by CCK-8 assay while the proliferation rate of cervical cancer cells was accelerated by overexpressed LINC01783 (Figure 2A). EdU experiment yielded the same results as CCK-8 assay (Figure 2B). Moreover, wound healing assay showed that the migration abilities of cervical cancer cells were reduced by downregulated LINC01783 and promoted by overexpressed LINC01783 (Figure 2C). Meanwhile, Transwell invasion assay showed that the invasion abilities of cervical cancer cells were reduced by downregulated LINC01783 and promoted by overexpressed LINC01783 (Figure 2D). LINC01783 overexpression mainly promoted S phase in C-33A cells. However, S phase was shortened in HeLa cells with LINC01783 knockdown (Figure 3A). The apoptosis rate of cervical cancer cells was increased by LINC01783 si, but inhibited by LINC01783 OE (Figure 3B). In summary, these results revealed that LINC01783 might exert regulatory effect on cell migration, proliferation, invasion, apoptosis and cycle progression of cervical cancer cells.
Subcellular Distribution of LINC01783
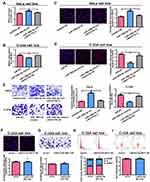
The biological functions of lncRNA depend on subcellular distribution.11,12 To verify the cellular location of LINC01783, cervical cancer cells were isolated into nuclear and cytoplasmic fractions, with U6 and GAPDH as controls, respectively. LINC01783 distribution was observed in the cytoplasmic fraction of C-33A and HeLa cells as shown by QRT-PCR (Figure 4A). A conclusion might be drawn that LINC01783 involved in the progression of cervical cancer via posttranscriptional regulation.
miR-199b-5p Is the Target of LINC01783
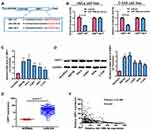
In view of the main distribution of LINC01783 in cytoplasmic fraction, it was postulated that LINC01783 might be a competitive endogenous RNA (ceRNA) in the progression of cervical cancer. According to qRT-PCR, in contrast to the trend of LINC01783 expression, miR-199b-5p expression was increased in cervical cancer cells (Figure 4B). The levels of miR-199b-5p are downregulated in cervical cancer tissues (Figure 4C) and negatively correlated to the expression of LINC01783 on TCGA dataset (Figure 4D). Starbase prediction revealed high matching of sequences in miR-199b-5p to LINC01783 3ʹUTR. pGL3-LINC01783 WT and pGL3-LINC01783 MUT were constructed based on such binding sequences (Figure 4E). Notable downregulation of luciferase activity was found in HeLa and C-33A cells post co-transfection with LINC01783 WT and miR-199b-5p mimics which was unchanged following transfection with LINC01783 MUT (Figure 4F). RIP analysis was used for describing whether LINC01783 involved in ribonucleoprotein complex containing RNAs. In QRT-PCR, LINC01783 was enriched in anti-Ago2 antibody relative to controls. miR-199b-5p obtained similar results (Figure 4G). It was suggested that miR-199b-5p could bind to LINC01783.
LINC01783 Regulates GBP1, a Target Gene of miR-199b-5p
Target genes of miR-199b-5p by bioinformatics prediction were screened out for the purpose of exploring the possible roles of miR-199b-5p in the progression of cervical cancer. Finally, GBP1 was selected for subsequent analyses. MiR-199b-5p mimics or miR-NCs were used for the co-transfection of constructed luciferase plasmids (pGL3-GBP1 WT and pGL3-GBP1 MUT) in HeLa and C-33A cells, respectively (Figure 5A). Luciferase activity of MUT reporter remained the same whereas WT reporter group was suppressed (Figure 5B). Based on the above results, GBP1 was a potential target of miR-199b-5p. Next, qRT-PCR was utilized for determining the GBP1 expression in cervical cancer cell lines. The RNA levels of GBP1 were notably enhanced in cervical cancer cells relative to those in HcerEpic cells (Figure 5C). In addition, the same result was obtained by Western blot analysis at the protein level (Figure 5D). The levels of GBP1 are upregulated in cervical cancer tissues (Figure 5E) and negatively correlated to the expression of miR-199b-5p on TCGA dataset (Figure 5F).
After adjusting LINC01783 or miR-199b-5p expression, the GBP1 expression in cervical cancer cells was detected for the purpose of clarifying whether LINC01783 regulated GBP1 expression by targeting miR-199b-5p. In HeLa cells, upregulation of GBP1 expression was observed following transfection with miR-199b-5p inhibitor. However, co-transfection with LINC01783 siRNA and miR-199b-5p inhibitor reversed this effect (Figure 6A and B). Moreover, inhibition of GBP1 expression was found following transfection with miR-199b-5p mimics in C-33A cells. However, co-transfection with LINC01783 overexpression plasmid and miR-199b-5p mimics reversed this effect (Figure 6C and D). Next, before determining GBP1 expression, LINC01783 WT overexpression plasmid and its mutant overexpression plasmid were used for the transfection of C-33A cells. Western blot and qRT-PCR revealed that GBP1 expression was upregulated in cervical cancer cells by overexpressed wild-type LINC01783, whereas the base pairing between LINC01783 and miR-199b-5p was not disrupted by mutant-type LINC01783 (Figure 6E and F). In brief, it was confirmed that LINC01783 showed a positive regulation of GBP1 expression through directly binding to miR-199b-5p.
LINC01783/miR-199b-5p Axis Regulates Behaviors of Cervical Cancer Cells
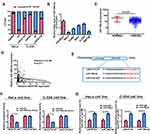
Subsequently, this study investigated whether miR-199b-5p affected the proliferative and invasive abilities of HeLa and C-33A cells. Firstly, qRT-PCR verified the transfection efficiency of miR-199b-5p inhibitor and mimics (Supplementary Figure 2C, D). Compared with controls, the proliferative and invasive abilities were considerably promoted by downregulating miR-199b-5p in HeLa cells; however, co-transfection with miR-199b-5p inhibitor and LINC01783 siRNA could partially reverse this effect (Figure 7A, C, E). Furthermore, the proliferation and invasion of C-33A cells were restrained by overexpression of miR-199b-5p, while LINC01783 overexpression could partially reverse this effect (Figure 7B, D, E). The proliferation, invasion, apoptosis and cell cycle progression were not affected by overexpression of mutant-type LINC01783 in C-33A cells (Figure 7F–I). Based on these results, LINC01783/miR-199b-5p/GBP1 axis presented with significant impact on the behavior regulation of cervical cancer cells.
Discussion
The cervical cancer rate is increasing worldwide. As shown by World Cancer Report (Geneva), the total annual rate of cervical cancer was 493,000 cases in 2002 and is expected to increase by nearly 42% (702,500 cases) as of 2020.13 It has been reported that LncRNA acts as a regulator of many cellular processes. It has also been identified that lncRNA dysregulation correlates with the disease development.14 Our data indicated highly expressed LINC01783 in cervical cancer cell line, and LINC01783 was shown to promote proliferation, migration, invasion and cycle, and inhibit apoptosis of HeLa and C-33A cells via function test.
A number of studies demonstrate that miRNA may act as a diagnostic biomarker and a tool to inhibit the protein translation.15–17 And, lncRNAs may bind to miRNA and control the function of miRNA for therapeutic purposes.18–20 A study suggests that MiR-199b-5 is a tumor-inhibiting factor in renal cell carcinoma.21 Another study indicates that miR-199b-5p restrains proliferative and invasive potentials of head and neck cancer cells.22 Our study found that miR-199b-5 bound to LINC01783 and showed low expression in cervical cancer cell lines.
Through bioinformatics prediction and dual luciferase reporter gene assay, we indicate that GBP1 is the potential target gene of miR-199b-5p. Previous studies showed that GBP1 promotes the progress of cancers. Ji X pointed out that overexpression of GBP1 predicts poor prognosis and promotes tumor growth in human glioblastoma multiforme.23 Li L found that GBP1 promotes lymph node metastasis in human esophageal squamous cell carcinoma.24 Our results showed a higher level of GBP1 in five cervical cancer cell lines compared with HcerEpic cells. Nevertheless, to further explore the expression of GBP1 in cervical cancer, the GBP1 Immunohistochemistry results of in situ expression of GBP1 protein in cervical cancer tissues should be complemented in further studies. Subsequently, we analyzed the expression of GBP1 in cervical cancer cells and the correlation between GBP1 levels and miR-199b-5p levels on TCGA dataset. Bioinformatics analysis showed that GBP1 is negatively correlated to miR-199b-5p. Then, Western blot assays showed that LINC01783 adjusted the expression of GBP1 which was a downstream target of miR-199b-5. Importantly, the results of biological function assays showed that miR-199b-5 mimics inhibited the proliferative and invasive abilities of cervical cancer cells whereas LINC01783 overexpression could partially reverse such effects of miR-199b-5 mimics on cervical cancer cells.
Conclusion
Taken together, LINC01783 was a ceRNA to regulate GBP1 expression by sponging miR-199b-5p, thereby regulating the progression of cervical cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Tsikouras P, Zervoudis S, Manav B, et al. Cervical cancer: screening, diagnosis and staging. J BUON. 2016;21(2):320–325.
2. Marret G, Borcoman E, Le Tourneau C. Pembrolizumab for the treatment of cervical cancer. Expert Opin Biol Ther. 2019;19:1–7.
3. de Freitas AC, Gurgel AP, Chagas BS, Coimbra EC, Do Amaral CM. Susceptibility to cervical cancer: an overview. Gynecol Oncol. 2012;126(2):304–311. doi:10.1016/j.ygyno.2012.03.047
4. Khan SR, Rockall AG, Barwick TD. Molecular imaging in cervical cancer. Q J Nucl Med Mol Imaging. 2016;60(2):77–92.
5. Yan Q, Tian Y, Hao F. Downregulation of lncRNA UCA1 inhibits proliferation and invasion of cervical cancer cells through miR-206 expression. Oncol Res. 2018. doi:10.3727/096504018X15185714083446
6. Wen Q, Liu Y, Lyu H, et al. Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int J Gynecol Cancer. 2017;27(6):1096–1108. doi:10.1097/IGC.0000000000001028
7. Xiao B, Huang Z, Zhou R, Zhang J, Yu B. The prognostic value of expression of the long noncoding RNA (lncRNA) Small Nucleolar RNA Host Gene 1 (SNHG1) in patients with solid malignant tumors: a systematic review and meta-analysis. Med Sci Monit. 2018;24:5462–5472. doi:10.12659/MSM.911687
8. Jing H, Qu X, Liu L, Xia H. A novel long noncoding RNA (lncRNA), LL22NC03-N64E9.1, promotes the proliferation of lung cancer cells and is a potential prognostic molecular biomarker for lung cancer. Med Sci Monit. 2018;24:4317–4323. doi:10.12659/MSM.908359
9. Man H, Bi W. Expression of a novel long noncoding RNA (lncRNA), GASL1, is downregulated in patients with intracranial aneurysms and regulates the proliferation of vascular smooth muscle cells in vitro. Med Sci Monit. 2019;25:1133–1139. doi:10.12659/MSM.912204
10. Misawa A, Takayama K, Urano T, Inoue S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J Biol Chem. 2016;291(34):17861–17880. doi:10.1074/jbc.M116.718536
11. Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23(7):1080–1087. doi:10.1261/rna.060814.117
12. Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41(9):761–772. doi:10.1016/j.tibs.2016.07.003
13. Bhat S, Kabekkodu SP, Noronha A, Satyamoorthy K. Biological implications and therapeutic significance of DNA methylation regulated genes in cervical cancer. Biochimie. 2016;121:298–311. doi:10.1016/j.biochi.2015.12.018
14. Li X, Wu Z, Fu X, Han W. lncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1–21. doi:10.1016/j.mrrev.2014.04.002
15. Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: challenges and prospects. Mol Diagn Ther. 2016;20(6):509–518. doi:10.1007/s40291-016-0221-4
16. Tiwari A, Mukherjee B, Dixit M. MicroRNA key to angiogenesis regulation: miRNA biology and therapy. Curr Cancer Drug Targets. 2018;18(3):266–277. doi:10.2174/1568009617666170630142725
17. Lin H, Ewing LE, Koturbash I, Gurley BJ, Miousse IR. MicroRNAs as biomarkers for liver injury: current knowledge, challenges and future prospects. Food Chem Toxicol. 2017;110:229–239. doi:10.1016/j.fct.2017.10.026
18. Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. doi:10.1093/bib/bbw053
19. Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med. 2018;16(1):264. doi:10.1186/s12967-018-1640-2
20. Cao MX, Jiang YP, Tang YL, Liang XH. The crosstalk between lncRNA and microRNA in cancer metastasis: orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 2017;8(7):12472–12483. doi:10.18632/oncotarget.13957
21. Lai Y, Quan J, Lin C, et al. miR-199b-5p serves as a tumor suppressor in renal cell carcinoma. Exp Ther Med. 2018;16(1):436–444. doi:10.3892/etm.2018.6151
22. Wang X, Li GH, Guo J, Sun Q. [miR-199b-5p inhibits proliferation and invasion of head and neck cancer cells]. Shanghai Kou Qiang Yi Xue. 2018;27(3):265–269.
23. Ji X, Zhu H, Dai X, et al. Overexpression of GBP1 predicts poor prognosis and promotes tumor growth in human glioblastoma multiforme. Cancer Biomark. 2019;25(3):275–290. doi:10.3233/CBM-171177
24. Li L, Ma G, Jing C, Liu Z. Guanylate-binding protein 1 (GBP1) promotes lymph node metastasis in human esophageal squamous cell carcinoma. Discov Med. 2015;20(112):369–378.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.