Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA LINC00466 Knockdown Inhibits Tongue Squamous Cell Carcinoma Malignancy by Targeting microRNA-493/HMGA2
Received 17 September 2020
Accepted for publication 23 November 2020
Published 22 December 2020 Volume 2020:12 Pages 13071—13084
DOI https://doi.org/10.2147/CMAR.S282625
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
This paper has been retracted.
Chao Hou, Yanli Dong, Bo Du
Department of Stomatology, Zaozhuang Municipal Hospital, Zaozhuang, Shandong 277100, People’s Republic of China
Correspondence: Bo Du
Department of Stomatology, Zaozhuang Municipal Hospital, 41 Zhonglongtou Road, Zaozhuang, Shandong 277100, People’s Republic of China
Email [email protected]
Purpose: Long intergenic non-protein-coding RNA 00466 (LINC00466) promotes lung adenocarcinoma progression. Nonetheless, the expression and precise roles of LINC00466 in tongue squamous cell carcinoma (TSCC) remains uncertain and warrant further investigation. Hence, the present study aimed to examine the LINC00466 effects on the aggressive TSCC cell characteristics and to elucidate the potential underlying mechanisms.
Methods: First, LINC00466 expression in TSCC was determined by reverse transcription-quantitative PCR. Subsequently, cell proliferation, apoptosis, migration, and invasion in vitro, as well as tumor growth in vivo were assessed to examine the LINC00466 effects on TSCC cells.
Results: LINC00466 was upregulated in TSCC. This upregulation was notably associated with shorter overall TSCC patient survival. In vitro experiments indicated that LINC00466 depletion suppressed TSCC cell proliferation, migration and invasion, and promoted apoptosis. An in vivo experiment revealed that LINC00466 downregulation attenuated TSCC tumor growth in vivo. Mechanistic analysis revealed that LINC00466 functions as a microRNA-493 (miR-493) molecular sponge, a miRNA that targets high-mobility group AT-hook 2 (HMGA2) mRNA. LINC00466 upregulated HMGA2 in TSCC cells, and this phenomenon was regulated by the miR-493 sponge. Rescue experiments revealed a decrease in the miR-493/HMGA2 axis output, partially reversing the effects of LINC00466 downregulation on aggressive TSCC cell behavior.
Conclusion: These findings demonstrate that LINC00466 promotes TSCC cell oncogenicity in vitro and in vivo by upregulating the miR-493/HMGA2 axis output. These results may provide a new perspective and new insight into the molecular mechanisms of TSCC.
Keywords: long intergenic non-protein-coding RNA 00466, tongue squamous cell carcinoma pathogenesis, targeted therapy
Expression of Concern for this paper has been published
Introduction
Tongue cancer is a prevalent type of head and neck carcinoma endangering public health worldwide.1 Tongue squamous cell carcinoma (TSCC) is the most common type of tongue cancer and exhibits characteristics, such as a high malignancy, unlimited growth, and active tissue infiltration that results in the malfunction of mastication, speech, and deglutition.2 Surgery plus radiotherapy, and neoadjuvant chemotherapy are the primary therapeutic methods for TSCC.3 Local relapse rates range from 18 to 76% among TSCC patients following first-line anticancer therapies, and local lymph node metastasis poses an enormous challenge for physicians treating TSCC patients.4 Despite recent advances in diagnostic and treatment methods, the clinical outcomes for TSCC patients have not improved significantly, and the 5-year overall survival rate is only 50%–60%.5 Even though TSCC pathogenesis has been investigated extensively over the past few decades, the detailed mechanisms and molecular events involved remain uncharacterized.6,7 Thus, it is critical to elucidate the mechanisms behind TSCC formation and progression. These findings may be useful for identifying potential targets for anticancer treatments.
Long non-coding RNAs (lncRNAs) are a heterogeneous family of non-protein-coding RNAs, typically over 200 nucleotides in length.8 Originally, lncRNAs were regarded as genomic ‘junk’ or ‘noise’ as they lack a protein-coding sequence.9 However, accumulating evidence suggests that lncRNAs are capable of interacting with RNA, proteins, and DNA. This interaction consequently regulates gene expression through different mechanisms.10–12 lncRNAs are involved in almost all physiological and pathological phenomena, including carcinogenesis and cancer progression.13 Lately, increasing numbers of studies have demonstrated the aberrant expression of lncRNAs in TSCC.14–17 lncRNAs possess both tumor-suppressive or cancer-promoting activities during TSCC initiation and progression. These processes include regulating the cell cycle, cell proliferation, apoptosis, metastasis, angiogenesis, and epithelial-mesenchymal transition.18–20 Accordingly, identifying lncRNAs contributing to TSCC pathogenesis is of utmost importance to discover novel diagnostic targets and treatments for this malignant tumor type.
A lncRNA, named long intergenic non-protein-coding RNA 00466 (LINC00466), was previously reported to facilitate tumor progression in lung adenocarcinoma.21 Nevertheless, the LINC00466 expression and precise roles in TSCC remain poorly studied. Therefore, the present study aimed to quantify LINC00466 expression in TSCC and to elucidate LINC00466’s clinical relevance in TSCC. Moreover, the LINC00466 effects on aggressive TSCC behavior were investigated and the possible LINC00466 mechanisms of action for TSCC progression were elucidated. We demonstrate for the first time that the LINC00466/miR-493/high-mobility group AT-hook 2 (HMGA2) pathway promotes TSCC progression. The pathway provides a promising set of targets for diagnosing, preventing, and/or treating TSCC.
Patients and Methods
Patients and Tissue Samples
The study was approved by the Zaozhuang Municipal Hospital Ethics Committee (ECZZMH-2014.0615) and was conducted following the World Medical Association Declaration of Helsinki. In addition, written informed consent was obtained from all study subjects. TSCC tissue samples and adjacent normal tissues were obtained from 53 patients with TSCC who had been admitted to Zaozhuang Municipal Hospital. None of these patients were treated with preoperative chemotherapy, radiotherapy, or other anti-tumor modalities. The clinicopathological characteristics of these patients were shown in Table 1. Follow-up was conducted for 5 years or until the patient’s death. Follow-up was executed by outpatient visits or by telephone. All the tissue samples collected were snap-frozen in liquid nitrogen and stored at −80°C.
 |
Table 1 The Clinicopathological Information of TSCC Patients |
Cell Lines and Cell Culture
Human TSCC cell lines (SCC-15 and CAL-27) were acquired from the American Type Culture Collection (ATCC) and were grown in Dulbecco’s modified Eagle’s medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific) and 1% penicillin/streptomycin mixture (Gibco; Thermo Fisher Scientific). The Minimum Essential Medium (Gibco; Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin/streptomycin mixture was used for cultivating normal gingival epithelial cells (ATCC® PCS-200-014™; ATCC). Cells were cultured at a constant temperature (37°C) supplied with 5% CO2.
Transient Transfection
Specific small interfering RNAs (siRNA) targeting LINC00466 (si-LINC00466#1 and si-LINC00466#2) and negative control (NC) siRNA (si-NC) were purchased from RiboBio Co. A miR-493 agomir (agomir-493) and miR-493 antagomir (antagomir-493) were synthesized by GenePharma Co., Ltd. and applied to increase or silence endogenous miR-493 expression, respectively. An NC agomir (agomir-NC) and NC antagomir (antagomir-NC) served as the controls for agomir-493 and antagomir-493, respectively. To restore HMGA2 expression, the full-length HMGA2 gene was amplified and inserted into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.), yielding the pcDNA3.1-HMGA2 plasmid (hereafter referred to as pc-HMGA2). Cells were transfected with plasmids or oligonucleotides using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Cellular Fractionation and Quantitative Reverse Transcription PCR (RT-qPCR)
Cellular fractionation was conducted using the Cytoplasmic & Nuclear RNA Purification kit (Norgen Biotek Corp.). Following separation of the nuclear and cytosolic fractions, RT-qPCR was carried out to assess LINC00466 expression inside TSCC cells.
RNA extraction was performed using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). To analyze HMGA2 mRNA and LINC00466 expression, complementary DNA was synthesized from each RNA sample using M-MLV reverse transcriptase (Promega Corporation). Following this, qPCR was performed using FastStart Universal SYBR-Green Master Mix (Roche Diagnostics). The relative HMGA2 mRNA and LINC00466 expression levels were normalized to GAPDH expression. To measure miR-493 expression, each RNA sample was reverse transcribed into complementary DNA using the miScript Reverse Transcription kit (Qiagen GmbH). The synthesized cDNA was then analyzed by qPCR via the miScript SYBR-Green PCR kit (Qiagen GmbH). The U6 small nuclear RNA served as an internal control for miR-493. The 2−ΔΔCq method22 was employed to calculate relative gene expression.
Cell Counting Kit-8 (CCK-8) Assay
Transfected cells were seeded in 96-well plates, with each well containing 2 × 103 cells in 200 μL of complete culture medium. Cellular proliferation was measured every 24 h: designated as day 0, 1, 2, and 3. A total of 10 µL of the CCK-8 solution (Dojindo Laboratories Co., Ltd.) was added to each well at each time point. Following a 2 h incubation at 37°C and 5% CO2, the absorbance at 450 nm was measured using a Sunrise Microplate Reader (Tecan Group, Ltd.).
Flow Cytometry
Apoptosis was evaluated using the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection kit (BioLegend). Transfected cells were harvested at 48 h post-transfection, and washed three times with ice-cold phosphate-buffered saline. The cells were then resuspended in 100 μL of flow cytometry binding buffer, and stained with 5 μL of Annexin V-FITC and 5 μL of propidium iodide solution. Following a 15 min incubation at room temperature in the dark, the proportion of apoptotic cells was assessed on a flow cytometer (FACScan™, BD Biosciences).
Transwell Migration and Invasion Assays
Transwell inserts with 8 µm pore size polycarbonate membranes (EMD Millipore) were used to determine the migratory and invasive abilities of TSCC cells. Migration assays were carried out using membranes not coated with Matrigel (BD Biosciences), whereas Matrigel-coated membranes (BD Biosciences) were used in the invasion assays. A cell suspension (200 µL) containing 5 × 104 transfected cells was added to the upper inserts. The bottom inserts contained 600 μL culture medium containing 10% of FBS. The non-migratory or non-invasive cells still inside the upper chamber were gently scraped off with a cotton swab following 24 h of incubation at 37°C. The migratory or invasive cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet solution (Beyotime Institute of Biotechnology, Inc.). Following extensive washing and air drying, the membranes were photographed under an inverted microscope (Olympus Corporation). Finally, the numbers of migratory and invasive cells were determined in 5 randomly selected visual fields per membrane.
In vivo Tumorigenesis Assay
To create stable LINC00466-deficient SCC-15 cells, a short hairpin RNA (shRNA) targeting LINC00466 (sh-LINC00466) and NC shRNA (sh-NC) were obtained from GenePharma Co., Ltd., and cloned into the pLKO.1 vector (Biosettia). This resulted in the creation of plasmids, pLKO.1-sh-LINC00466, and pLKO.1-sh-NC. HEK293T cells were transduced with either pLKO.1-sh-LINC00466 or pLKO.1-sh-NC in the presence of psPAX2 and pMD2.G. Lentiviruses expressing either sh-LINC00466 or sh-NC were collected 2 days after transduction and subsequently introduced to SCC-15 cells. Puromycin (5 μg/mL; Sigma-Aldrich; Merck KGaA) was used to select stable LINC00466-deficient cells.
All animal experiments were approved by the Committee on Ethics of Animal Experiments at Zaozhuang Municipal Hospital (ECAZZMH-2018.0902) and performed in strict accordance with NIH guidelines for the care and use of laboratory animals. For this study, 4- to 6-week-old male BALB/c nude mice (weight, 20 g) were acquired from Shanghai SLAC Laboratory Animal Co., Ltd. SCC-15 cells stably expressing either sh-LINC00466 or sh-NC were injected subcutaneously into the dorsal flanks of the mice. Tumor size was measured using calipers every 3 days. Tumor xenografts volumes were calculated using the following formula: Volume = 0.5 × length × width2. All the mice were euthanized employing cervical dislocation 30 days after cell injection. No injectable anesthetics were used in the present study. Tumor xenografts were collected, photographed, and weighed. Tumors were then imaged and stored for RT-qPCR and Western blot analysis.
Bioinformatics Analysis
microRNAs (miRNAs or miRs) binding to the lncRNA was predicted from the starBase 3.0 database (http://starbase.sysu.edu.cn/).
Luciferase Reporter Assay
LINC00466 fragments containing either the wild-type (wt) miR-493-binding site or a mutant (mut) binding site were amplified by GenePharma Co., Ltd., and cloned into the psiCHECK2 luciferase reporter vector (Promega Corporation). This process yielded plasmids LINC00466-wt and LINC00466-mut. The wt or mut luciferase reporter plasmid was co-transfected with either agomir-493 or agomir-NC into TSCC cells using Lipofectamine 2000. Firefly and Renilla luciferase activities were determined by the Dual-Luciferase Reporter assay (Promega Corporation) following 48 h of cell culture at 37°C. Renilla luciferase activity was normalized to Firefly luciferase activity.
RNA Immunoprecipitation (RIP) Assay
The RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (EMD Millipore). TSCC cells were lysed in RIP lysis buffer at 4°C, centrifuged to obtain a debris-free cell lysate, and subjected to overnight incubation at 4°C with magnetic beads conjugated with an anti-Argonaute 2 (AGO2) or control IgG antibody (both from EMD Millipore). Magnetic beads were collected and incubated with proteinase K at 55°C for 30 min to digest proteins. Finally, the immunoprecipitated RNA was extracted and analyzed by RT-qPCR.
Western Blot Analysis
Cells were lysed with ice-cold RIPA lysis buffer (Beyotime Institute of Biotechnology, Inc.). The extracted total protein was quantified using a Bicinchoninic Acid Assay kit (Beyotime Institute of Biotechnology, Inc.). Equal amounts of protein were separated on a 10%SDS-PAGE gel and transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% fat-free milk at room temperature for 2 h. Following overnight incubation at 4°C with primary antibodies, the membranes were rinsed with Tris-buffered saline containing 0.1% of Tween-20 and incubated with a horseradish peroxidase-conjugated secondary antibody (cat. no. ab205718; Abcam) for 2 h at room temperature. Following an additional wash with Tris-buffered saline containing 0.1% Tween-20, chemiluminescence quantification and signal development were performed using the Enhanced Chemiluminescence Reagent (Bio-Rad Laboratories, Inc.). The primary antibodies anti-HMGA2 antibody (cat. no. ab207301; Abcam) and anti-GAPDH antibody (cat. no. ab181602; Abcam) were used at a 1:1000 dilution.
Statistical Analysis
All data are presented as the mean ± SD. Comparisons between 2 groups were conducted using a paired and unpaired Student’s t-test. One-way analysis of variance with Tukey’s post hoc test was used out to assess the differences among multiple groups. The overall patient survival with TSCC was analyzed via the Kaplan–Meier method, and the Log rank test was performed for univariate analysis. Correlations between the LINC00466, miR-493, and HMGA2 expression levels in the 53 TSCC tissue samples were calculated using Spearman’s rank-order correlation. Statistical significance was assumed when a P-value was < 0.05.
Results
LINC00466 Upregulation in TSCC Patients is Associated with a Poor Prognosis
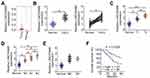
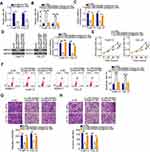
To determine the function of LINC00466 in TSCC, its expression in Head and Neck squamous cell carcinoma (HNSC) was first analyzed employing The Cancer Genome Atlas (TCGA) dataset. LINC00466 was upregulated in tumor tissues in comparison with that in normal tissues (Figure 1A). In addition, LINC00466 expression in 53 pairs of TSCC tissue samples and adjacent normal tissues were detected by RT-qPCR. Compared with adjacent normal tissues, LINC00466 expression was higher in TSCC tissues (Figure 1B). Subsequently, the clinical relevance of LINC00466 in TSCC was determined. A high LINC00466 expression was significantly correlated with tumor size (Figure 1C) and lymph node metastasis (Figure 1D) but not related with distant metastasis (Figure 1E) in patients with TSCC. Study subjects were binned into a high-LINC00466 expression group or a low-LINC00466 expression group, based on the LINC00466 expression median value among all TSCC tissue samples. Patients in the high-LINC00466 expression group exhibited a shorter overall survival than the patients in the low-LINC00466 expression group (Figure 1F; P = 0.0305). These results indicated that LINC00466 upregulated in TSCC and may be important for TSCC tumor progression.
LINC00466 Depletion Weakens the TSCC Malignant Phenotype in vitro
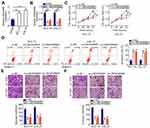
LINC00466 expression was quantified by RT-qPCR in TSCC cell lines (SCC-15 and CAL-27) and normal gingival epithelial cells. The results revealed that LINC00466 expression was higher in TSCC cell lines than in normal gingival epithelial cells (Figure 2A). SCC-15 and CAL-27 is widely used in functional experiments regarding TSCC and manifested overexpressed LINC00466 expression; thus, the two cell lines were selected for use in subsequent loss-of-function assay. To further analyze the LINC00466 effects on the biological characteristics of TSCC, LINC00466 reducing siRNAs (si-LINC00466#1 and si-LINC00466#2) were separately transfected into SCC-15 and CAL-27 cells to knock down LINC00466 expression. RT-qPCR was used to assess transfection efficiency (Figure 2B).
CCK-8 assay was conducted to assess the effects of LINC00466 on TSCC cell proliferation. Transfection with si-LINC00466 resulted in decreased proliferation for SCC-15 and CAL-27 cells (Figure 2C). Flow cytometry showed the proportion of apoptotic SCC-15 and CAL-27 cells was substantially increased when was LINC00466 knocked down (Figure 2D). Furthermore, transwell migration and invasion assays suggested that LINC00466 knockdown significantly suppressed SCC-15 and CAL-27 cell migratory (Figure 2E) and invasive (Figure 2F) abilities. The above-mentioned results indicate that LINC00466 may function as an oncogenic lncRNA, promoting TSCC progression.
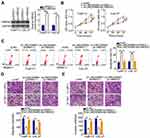
LINC00466 Functions as a Molecular Sponge of miR-493 in TSCC Cells
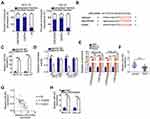
lncRNAs may serve as competing endogenous RNAs (ceRNAs) by directly interacting with and sequestering miRNAs.23 To elucidate the mechanisms through which LINC00466 affects TSCC progression, we determined the location of LINC00466 in SCC-15 and CAL-27 cells. LINC00466 was mainly located in the cytoplasm of the SCC-15 and CAL-27 cells (Figure 3A). We hypothesized that LINC00466 may function as a ceRNA, sequestering certain miRNAs in TSCC cells. The online database, starBase 3.0, was used to search for predicted LINC00466 target miRNAs. LINC00466 contained a putative miR-493–binding site (Figure 3B).
A luciferase reporter assay was performed to confirm the complementary base pairing between miR-493 and LINC00466. First, the transfection efficiency of agomir-493 was validated by RT-qPCR (Figure 3C). The luciferase reporter assay showed the luciferase activity in the SCC-15 and CAL-27 cells decreased by co-transfection with agomir-493 and LINC00466-wt. However, miR-493 overexpression did not affect LINC00466-mut luciferase activity (Figure 3D). RIP assay revealed miR-493 and LINC00466 were enriched following immunoprecipitation with anti-AGO2 antibody (compared to IgG) in SCC-15 and CAL-27 cell lysates (Figure 3E). This indicated a direct interaction between miR-493 and LINC00466 in TSCC cells.
miR-493 expression in the 53 pairs of TSCC tissue samples and adjacent normal tissues was also measured by RT-qPCR. The results indicated that miR-493 expression was lower in TSCC tissues compared to adjacent normal tissues (Figure 3F). In addition, miR-493 expression inversely correlated with LINC00466 expression in the 53 TSCC tissue samples (Figure 3G; rs = −0.6250, P < 0.0001). Subsequently, RT-qPCR was performed to examine the LINC00466 regulatory effects on miR-493 expression in TSCC cells. miR-493 expression was upregulated in SCC-15 and CAL-27 cells after LINC00466 expression was silenced (Figure 3H). Overall, these findings suggested that LINC00466 directly targets miR-493 and functions as a miR-493 molecular sponge in TSCC cells.
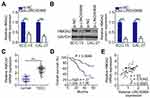
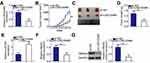
LINC00466 Positively Regulates HMGA2 Expression in TSCC Cells
Given that HMGA2 is a direct target gene of miR-493 in TSCC cells,24 we sought to determine whether LINC00466 is involved in regulating HMGA2 expression. SCC-15 and CAL-27 cells were transfected with either si-LINC00466 or si-NC. The transfected cells were then subjected to RT-qPCR and Western blot to determine HMGA2 mRNA and protein expression, respectively. Inhibiting LINC00466 expression downregulated HMGA2 expression in SCC-15 and CAL-27 cells at the mRNA (Figure 4A) and protein level (Figure 4B). Additionally, the HMGA2 mRNA level was quantified in the 53 pairs of TSCC tissue samples. HMGA2 mRNA level was higher in the TSCC tissues when compared to the adjacent normal tissues (Figure 4C). TSCC patients with high HMGA2 expression manifested a shorter overall survival in contrast to those with low HMGA2 expression (Figure 4D; P = 0.0046). Furthermore, a positive correlation between HMGA2 mRNA and LINC00466 expression levels in the 53 TSCC tissue samples (Figure 4E; rs = 0.5342, P < 0.0001). Thus, these results indicate that LINC00466 functions as a ceRNA on miR-493, and increasing HMGA2 expression in TSCC cells.
The LINC00466/miR-493/HMGA2 Axis Promotes TSCC Progression
To examine whether the miR-493/HMGA2 axis is indispensable for LINC00466-mediated TSCC progression, a series of rescue assays were performed. Before the assays, the efficiency of antagomir-493 was analyzed by RT-qPCR. The results revealed that transfection with antagomir-493 notably silenced miR-493 expression in both SCC-15 and CAL-27 cells (Figure 5A). Subsequently, either antagomir-493 or antagomir-NC was co-transfected with si-LINC00466 in SCC-15 and CAL-27 cells. The increase in miR-493 levels induced in SCC-15 and CAL-27 cells by LINC00466 knockdown was attenuated by co-transfection with antagomir-493, as shown by the RT-qPCR data (Figure 5B). In addition, the si-LINC00466 effects on HMGA2 mRNA (Figure 5C) and protein (Figure 5D) expression were partly reversed by antagomir-493 transfection into SCC-15 and CAL-27 cells. In functional assays, miR-493 silencing reversed the si-LINC00466 effects on proliferation (Figure 5E), apoptosis (Figure 5F), migration (Figure 5G), and invasiveness (Figure 5H) in SCC-15 and CAL-27 cells.
An HMGA2 overexpression plasmid (pc-HMGA2) was constructed, and its efficiency was examined by Western blot analysis (Figure 6A). The pc-HMGA2 or empty pcDNA3.1 vector was transfected into SCC-15 and CAL-27 cells in the presence of si-LINC00466. HMGA2 expression restoration attenuated the LINC00466 knockdown effects in SCC-15 and CAL-27 cell proliferation (Figure 6B), apoptosis (Figure 6C), migration (Figure 6D), and invasion (Figure 6E). Overall, LINC00466 contributed to TSCC progression by upregulating the miR-493/HMGA2 axis.
LINC00466 Depletion Suppresses TSCC Cell Growth in vivo
A tumorigenesis experiment was conducted to examine the LINC00466 effects on TSCC tumor growth in vivo. First, LINC00466 expression was determined in SCC-15 cells stably transfected with either sh-LINC00466 or sh-NC. The RT-qPCR results suggested that LINC00466 expression was very low in SCC-15 cells infected with the sh-LINC00466 lentivirus (Figure 7A). SCC-15 cells stably expressing either sh-LINC00466 or sh-NC were injected subcutaneously into the dorsal flanks of nude mice. The tumor volume (Figure 7B and C) and weight (Figure 7D) were smaller in the sh-LINC00466 group than in the sh-NC group. The RT-qPCR results showed tumor miR-493 expression was significantly higher in the sh-LINC00466 group compared to the sh-NC group (Figure 7E). The HMGA2 mRNA (Figure 7F) and protein levels (Figure 7G) in tumor xenografts derived from sh-LINC00466-transfected SCC-15 cells were substantially lower compared to sh-NC tumor xenografts. Overall, these findings suggested that LINC00466 knockdown suppressed TSCC growth in vivo by regulating the miR-493/HMGA2 axis.
Discussion
A growing body of evidence suggests that numerous lncRNAs are aberrantly expressed in TSCC.14,20,25 lncRNAs are major molecular regulators implicated in modulating multiple malignant characteristics during TSCC initiation and progression.26 Consequently, the in-depth elucidation of TSCC-associated lncRNA functions in tumor progression may provide novel diagnostic and therapeutic perspectives for managing TSCC. In the present study, the LINC00466 expression in TSCC was measured and its clinical relevance was determined. Additionally, the LINC00466 effects on the aggressive TSCC cell phenotype were investigated in vitro and in vivo. The mechanisms behind the LINC00466 cancer-promoting effects on TSCC progression were also clarified.
LINC00466 is upregulated in lung adenocarcinoma.21 In terms of its function, LINC00466 downregulation inhibits lung adenocarcinoma cell proliferation, migration, and invasion, as well as induced apoptosis in vitro, and reduced tumorigenicity in vivo.21 To the best of our knowledge, the LINC00466 expression profiles and roles in TSCC have not yet been reported. In the present study, RT-qPCR was performed to assess LINC00466 expression in TSCC tumors and cell lines. We revealed that LINC00466 was upregulated in both sources of TSCC samples and cell lines. TSCC patients with high-LINC00466 expression exhibited adverse clinicopathological characteristics and a shorter overall survival than patients with low-LINC00466 expression. In vitro experiments indicated that LINC00466 knockdown suppressed TSCC cell proliferation, migration, and invasion in vitro. In addition, LINC00466 knockdown promoted TSCC cell apoptosis. In vivo experiments revealed that LINC00466 knockdown attenuated TSCC tumor growth in vivo. These findings suggest LINC00466 as a prognostic biomarker and therapeutic target for TSCC.
Recent research indicates the existence of ceRNA interaction networks, in which lncRNAs function as molecular sponges that bind to miRNAs and consequently sequester the miRNA targets.27,28 Therefore, we investigated the possibility that LINC00466 was a molecular sponge that promotes TSCC growth by regulating downstream molecular events underlying tumor-promoting activities. Cellular fractionation indicated that LINC00466 was mainly located in the cytoplasm of TSCC cells, suggesting LINC00466 functions as a miRNA sponge. Bioinformatics analysis revealed a putative miR-493-binding site within LINC00466. Experiments conducted confirmed bioinformatics results. The luciferase reporter and RIP assays revealed that LINC00466 can specifically bind and interact with miR-493 in TSCC cells. Additionally, miR-493 expression was markedly downregulated in TSCC tissues when compared to adjacent normal tissues. These findings are consistent with observations from a previous study.24 Moreover, an inverse correlation between LINC00466 and miR-493 expression levels in 53 TSCC tissue samples was identified by Spearman correlation. Further experiments revealed that LINC00466 knockdown increased miR-493 expression and decreased HMGA2 expression in TSCC cells. Hence, the role of LINC00466 in TSCC malignancy can be partly explained by a ceRNA mechanism. Our study, for the first time, identified a novel ceRNA pathway in ceRNA involving involving LINC00466, miR-493, and HMGA2 mRNA.
miR-493 dysregulation in a variety of human cancers has been widely reported.29–34 miR-493 expression is low in TSCC and is significantly associated with adverse clinical characteristics and poor clinical outcomes.24 Functional experiments have confirmed miR-493 as an antioncogenic miRNA important for TSCC progression.24 Mechanistic experiments have identified HMGA2 mRNA as a direct miR-493 target in TSCC cells.24 HMGA2, a member of the high-mobility group A protein family, is upregulated and functions as an oncogenic protein in TSCC.35,36 In the present study, HMGA2 was also upregulated in TSCC. TSCC patients with high HMGA2 expression had a shorter overall survival than those with low HMGA2 expression. LINC00466 positively regulates HMGA2 expression in TSCC cells and this effect link to miR-493 sequestration. Furthermore, the expression of HMGA2 was positively correlated with LINC00466 levels in TSCC tissues. Rescue experiments indicated a decrease in the miR-493/HMGA2 axis output, partially reversing the LINC00466 downregulated effects on TSCC’s aggressive behavior. These results support the notion that the LINC00466/miR-493/HMGA2 pathway is functional and involved in TSCC pathogenesis, which might be an effective therapeutic route for TSCC.
Conclusion
In conclusion, LINC00466 upregulation is associated with a poor TSCC patient prognosis. LINC00466 promotes TSCC progression and plays a critical part in regulating tumor cell proliferation, apoptosis, migration, invasion, and ultimately tumor growth. LINC00466 functions as a miR-493 sponge and consequently attenuates the negative miR-493 regulatory effects on HMGA2 expression and promoting TSCC malignancy. The findings in this present study reveal crucial LINC00466 functions in TSCC cell oncogenicity and suggests possible LINC00466 applications in anticancer therapies.
Ethics Approval
The study protocol was approved by the Zaozhuang Municipal Hospital Ethics Committee (ECZZMH-2014.0615) and was conducted following the World Medical Association Declaration of Helsinki. All animal experiments were approved by the Committee on Ethics of Animal Experiments at Zaozhuang Municipal Hospital (ECAZZMH-2018.0902) and performed in strict accordance with NIH guidelines for the care and use of laboratory animals.
Consent for Publication
Not applicable.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Yang TL, Wang CP, Ko JY, Lin CF, Lou PJ. Association of tumor satellite distance with prognosis and contralateral neck recurrence of tongue squamous cell carcinoma. Head Neck. 2008;30(5):631–638. doi:10.1002/hed.20758
2. Ramqvist T, Grün N, Dalianis T. Human papillomavirus and tonsillar and base of tongue cancer. Viruses. 2015;7(3):1332–1343. doi:10.3390/v7031332
3. Schwam ZG, Judson BL. Improved prognosis for patients with oral cavity squamous cell carcinoma: analysis of the national cancer database 1998–2006. Oral Oncol. 2016;52:45–51. doi:10.1016/j.oraloncology.2015.10.012
4. Warnakulasuriya S. Living with oral cancer: epidemiology with particular reference to prevalence and life-style changes that influence survival. Oral Oncol. 2010;46(6):407–410. doi:10.1016/j.oraloncology.2010.02.015
5. Safi AF, Grandoch A, Nickenig HJ, Zöller JE, Kreppel M. The importance of lymph node ratio for locoregional recurrence of squamous cell carcinoma of the tongue. J Cranio-Maxillo-Facial Surgery. 2017;45(7):1058–1061. doi:10.1016/j.jcms.2017.04.008
6. Knopf A, Lempart J, Bas M, Slotta-Huspenina J, Mansour N, Fritsche MK. Oncogenes and tumor suppressor genes in squamous cell carcinoma of the tongue in young patients. Oncotarget. 2015;6(5):3443–3451. doi:10.18632/oncotarget.2850
7. Regezi JA, Dekker NP, McMillan A, et al. p53, p21, Rb, and MDM2 proteins in tongue carcinoma from patients < 35 versus > 75 years. Oral Oncol. 1999;35(4):379–383.
8. Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi:10.4161/rna.20481
9. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
10. Xin Y, Li Z, Shen J, Chan MT, Wu WK. CCAT1: A pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016;49(3):255–260. doi:10.1111/cpr.12252
11. Yu Y, Yang J, Li Q, Xu B, Lian Y, Miao L. LINC00152: A pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50:4.
12. Zhao J, Zhang C, Gao Z, Wu H, Gu R, Jiang R. Long non-coding RNA ASBEL promotes osteosarcoma cell proliferation, migration, and invasion by regulating microRNA-21. J Cell Biochem. 2018;119(8):6461–6469. doi:10.1002/jcb.26671
13. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
14. Gao W, Chan JY, Wong TS. Long non-coding RNA deregulation in tongue squamous cell carcinoma. Biomed Res Int. 2014;2014:405860. doi:10.1155/2014/405860
15. Huang W, Cui X, Chen J, et al. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget. 2016;7(38):62520–62532. doi:10.18632/oncotarget.11528
16. Li ZQ, Zou R, Ouyang KX, Ai WJ. An in vitro study of the long non-coding RNA TUG1 in tongue squamous cell carcinoma. J Oral Pathol Med. 2017;46(10):956–960.
17. Wang ZY, Hu M, Dai MH, et al. Upregulation of the long non-coding RNA AFAP1-AS1 affects the proliferation, invasion and survival of tongue squamous cell carcinoma via the Wnt/beta-catenin signaling pathway. Mol Cancer. 2018;17(1):3. doi:10.1186/s12943-017-0752-2
18. Yu J, Liu Y, Gong Z, et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8(10):16621–16632. doi:10.18632/oncotarget.14200
19. Yu J, Liu Y, Guo C, et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8(4):523–530. doi:10.7150/jca.17510
20. Zhang H, Zhao L, Wang YX, Xi M, Liu SL, Luo LL. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumour Biol. 2015;36(11):8805–8809. doi:10.1007/s13277-015-3645-2
21. Ma T, Hu Y, Guo Y, Yan B. Tumor-promoting activity of long noncoding RNA LINC00466 in lung adenocarcinoma via miR-144-Regulated HOXA10 axis. Am J Pathol. 2019;189(11):2154–2170. doi:10.1016/j.ajpath.2019.06.014
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
23. Ye Y, Shen A, Liu A. Long non-coding RNA H19 and cancer: A competing endogenous RNA. Bull Cancer. 2019;106(12):1152–1159. doi:10.1016/j.bulcan.2019.08.011
24. Jiao D, Liu Y, Tian Z. microRNA-493 inhibits tongue squamous cell carcinoma oncogenicity via directly targeting HMGA2. Onco Targets Ther. 2019;12:6947–6959. doi:10.2147/OTT.S210567
25. Fang Z, Zhang S, Wang Y, et al. Long non-coding RNA MALAT-1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins. BMC Cancer. 2016;16:706. doi:10.1186/s12885-016-2735-x
26. Wang J, Li L, Wu K, et al. Knockdown of long noncoding RNA urothelial cancer-associated 1 enhances cisplatin chemosensitivity in tongue squamous cell carcinoma cells. Die Pharmazie. 2016;71(10):598–602.
27. Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:5. doi:10.3390/ijms19051310
28. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi:10.1002/jcp.27941
29. Zhou W, Zhang C, Jiang H, Zhang Z, Xie L, He X. MiR-493 suppresses the proliferation and invasion of gastric cancer cells by targeting RhoC. Iran J Basic Med Sci. 2015;18(10):1027–1033.
30. Cui A, Jin Z, Gao Z, et al. Downregulation of miR-493 promoted melanoma proliferation by suppressing IRS4 expression. Tumour Biol. 2017;39(5):1010428317701640. doi:10.1177/1010428317701640
31. Xu Y, Ge K, Lu J, Huang J, Wei W, Huang Q. MicroRNA-493 suppresses hepatocellular carcinoma tumorigenesis through down-regulation of anthrax toxin receptor 1 (ANTXR1) and R-spondin 2 (RSPO2). Biomed Pharmacother. 2017;93:334–343. doi:10.1016/j.biopha.2017.06.047
32. Zhi D, Zhao X, Dong M, Yan C. miR-493 inhibits proliferation and invasion in pancreatic cancer cells and inversely regulated hERG1 expression. Oncol Lett. 2017;14(6):7398–7404.
33. Ueno K, Hirata H, Majid S, et al. Tumor suppressor microRNA-493 decreases cell motility and migration ability in human bladder cancer cells by downregulating RhoC and FZD4. Mol Cancer Ther. 2012;11(1):244–253. doi:10.1158/1535-7163.MCT-11-0592
34. Gu Y, Cheng Y, Song Y, et al. MicroRNA-493 suppresses tumor growth, invasion and metastasis of lung cancer by regulating E2F1. PLoS One. 2014;9(8):e102602. doi:10.1371/journal.pone.0102602
35. Zhao XP, Zhang H, Jiao JY, Tang DX, Wu YL, Pan CB. Overexpression of HMGA2 promotes tongue cancer metastasis through EMT pathway. J Transl Med. 2016;14:26. doi:10.1186/s12967-016-0777-0
36. Zhang H, Tang Z, Deng C, et al. HMGA2 is associated with the aggressiveness of tongue squamous cell carcinoma. Oral Dis. 2017;23(2):255–264. doi:10.1111/odi.12608
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.