Back to Journals » OncoTargets and Therapy » Volume 14
Long Non-Coding RNA LINC00313 Accelerates Cervical Carcinoma Progression by miR-4677-3p/CDK6 Axis
Authors Zhai Y, Liu Y, Wang Z, Wang W, Zhou J, Lu J
Received 28 May 2020
Accepted for publication 4 February 2021
Published 29 March 2021 Volume 2021:14 Pages 2213—2226
DOI https://doi.org/10.2147/OTT.S265007
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Arseniy Yuzhalin
Yongning Zhai,1,* Yang Liu,1,* Zhen Wang,1 Wei Wang,2 Juan Zhou,1 Jingyuan Lu3
1Department of Gynecology, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, Jiangsu Province, People’s Republic of China; 2Department of Pathology, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, Jiangsu Province, People’s Republic of China; 3Department of Radiological Intervention, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, Jiangsu Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jingyuan Lu
Department of Radiological Intervention, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, Jiangsu Province, People’s Republic of China
Email [email protected]
Juan Zhou
Department of Gynecology, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, Jiangsu Province, People’s Republic of China
Email [email protected]
Background: Cervical cancer is one of the most common gynecologic tumors. Evidence is accumulating that long non-coding RNAs participate in the pathogenesis of cancers, but the expression and role of lncRNA LINC00313 in cervical carcinoma is not reported.
Methods: We measured the expression levels of LINC00313 in clinical samples of cervical carcinoma and investigated the function of LINC00313 in the regulation of proliferation, metastasis, and EMT. Luciferase reporter assay was employed to explore the molecular regulation process of LINC00313.
Results: Our data showed that the levels of LINC00313 in cervical carcinoma tissues and cells were significantly up-regulated. Functionally, LINC00313 accelerated the progression, migration, and EMT of SiHa and Hela cells. Luciferase reporter assay confirmed that miR-4677-3p/CDK6 regulatory axis is the direct downstream of LINC00313. Functional gain- and loss-of-function strategies further showed that LINC00313 induced the up-regulation of CDK6 expression through competitive binding with miR-4677-3p, leading to promote the progression of cervical carcinoma.
Conclusion: Our results demonstrated that LINC00313 accelerated the progression of cervical cancer through the miR-4677-3p/CDK6 regulatory axis. LncRNA LINC00313 may serve as a potential target for the diagnosis and treatment of cervical carcinoma.
Keywords: cervical carcinoma, LINC00313, miR-4677-3p, CDK6
Introduction
Cervical carcinoma is the second most common in gynecologic tumors and the leading cause of death among women in developing countries.1 Due to the difficulty of achieving the early screening of cervical cancer in time, the higher prevalence rate occurs in developing countries, especially in low-income countries.2 Although early screening reduces the mortality of cervical carcinoma, invasion, metastasis, and recurrence were the leading cause of death.3 Consequently, it is urgent to find novel therapeutic targets and prognostic biomarkers to improve the survival rate of cervical carcinoma patients.
Long non-coding RNA (lncRNA) is a kind of non-coding RNA whose length is larger than 200 nt.4 Increasing articles have reported that lncRNAs regulate the expression of downstream genes, and then participate in the occurrence and development of tumors.5–7 For example, the up-regulation of lncRNA MALAT1 in cervical carcinoma tissues significantly affects the tumor size, TNM stage, and lymph node metastasis.8,9 Also, the negative regulation of GAS5 in cervical carcinoma was also significantly correlated with the progression.10,11 In recent studies, the expression of LINC00313 was found to decrease the expression of ALX4 in thyroid carcinoma.12 However, the role of LINC00313 in the pathogenesis of cervical carcinoma is not explored.
In this study, we firstly reported that LINC00313 was overexpressed in cervical carcinoma tissues and cells. Subsequent functional studies showed that the knockdown of LINC00313 significantly inhibited the progression, migration, and EMT of cervical cancer in vitro. Furthermore, we predicted and confirmed that miR-4677-3p/CDK6 regulatory axis was the direct downstream of LINC00313. The silencing effect of LINC00313 on the cervical cancer progression was significantly blocked by the knockdown of miR-4677-3p, which could be further reversed by the downregulation of CDK6. Collectively, our results demonstrated that LINC00313 accelerates the progression of cervical cancer by inhibiting the miR-4677-3p/CDK6 axis. LncRNA LINC00313 may serve as a potential target for the diagnosis and treatment of cervical carcinoma.
Methods
Clinical Specimens
Fresh cervical cancer tissues were acquired from 50 patients at Nanjing Maternity and Child Health Care Hospital from June 2017 to December 2019. Moreover, the healthy cervical tissues at the edge of the cervical tumor were obtained as the paracancerous tissue (control). Subsequently, the specimens were quickly snap-frozen in liquid nitrogen and transferred to −80°C for preservation. All the patients signed informed consent, and the hospital ethical committee approved this study (Permit number:2019-KY-007).
To determine whether a patient had a low or high expression of LINC00313, miR-4677-3p and CDK6. We analyzed the slices of the tumor samples by Immunohistochemistry or in situ hybridization (ISH). ISH probes (all purchased from GenePharma Inc., Shanghai, China) were employed for the expression detection of Lnc-RNA LINC00313 and miR-4677-3p. And, antibodies (ab151247, Abcam) were used for the expression detection of CDK6. The sample was identified as positive if more than 50% of total cells were positive.
Cell Treatment
H8 (human cervical epithelium cell lines) and four types of cervical cancer cell lines, including CaSki, SiHa, Hela, and C33A cells, were employed. All the cell lines were obtained from ATCC and cultured according to their recommendations. miRNA mimics/inhibitors and their corresponding control were commercially obtained from GenePharm Co. Ltd. (Shanghai, China). For gene overexpression, the full-length gene of LINC00313 was cloned into the pcDNA3.1 (+) vector (Sigma, MO, USA). In the experiment of gene knockdown, short hairpin RNAs (shRNAs) targeting the LINC00313 and CDK6 were commercially synthesized and cloned into pLKO.1 vector (Sigma, MO, USA). When the cell density reached 70–80%, plasmids, and miRNA mimics or inhibitors were transfected by Lipofectamine 3000 (ThermoFisher, Shanghai, China). Under the specified plural number of infection and treatment time, these vectors and miRNA mimics or inhibitors have no side effects, including affecting the adherence, shape, or activity of cervical cells mentioned above.
Animal Experiments
All animal experiments were conducted following national and international guidelines and approved by the Animal Care and Utilization institutions Review Committee of Nanjing Maternity and Child Health Care Hospital. 6-week-old male Balb/c nude mice were placed in the absence of specific pathogen (SPF) and randomly divided into sh-NC + NC inhibitor group, sh-LINC00313 + NC inhibitor group, sh-LINC00313 + miR-4677-3p inhibitor group, and sh-LINC00313 + miR-4677-3p inhibitor + sh-CDK6 group (n=3 in each group). For tumorigenicity in vivo, 6×106 transfected Hela cells were subcutaneously injected into the right side of mice. The tumor volume was measured every week and calculated according to the formula: 0.5 × length × width2. All mice were euthanatized 35 days later, and the tumors were surgically removed, measured, and stored in liquid nitrogen for further experiments.
Quantitative Real-Time PCR
Cells were treated according to the experimental group. When the density reached 90%, the RNA extraction was performed by TRIzol reagent (Beyotime, Nantong, China). According to the instructions, the extracted total RNA was reverse transcribed to cDNA by PrimeScript 1st Strand cDNA synthesis kit (Takara, Otsu, Japan). After the implementation of real-time reverse-transcription polymerase chain reaction, the CT value of the target gene expression was obtained compared with the control group. U6 was used as the internal control gene. The relative quantitative analysis was carried out by the 2–ΔΔCT method. The probes were synthesized as followed: U6-F: 5ʹ-CCG CCC GCC GCC AGG CCCC-3ʹ; U6-R: 5ʹ-ATA TGG AAC GCT TCA CGA ATT-3ʹ; LINC00313-F: 5ʹ- GGA AGC ACT TAG ACC CTG CC-3ʹ; LINC00313-R: 5ʹ- GCC GCT GTT GGT TTC ATC TC-3ʹ; miR-4677-3p-F: 5ʹ-CTG TGA GAC CAA AGA ACT ACT CGC-3ʹ; miR-4677-3p-R: 5ʹ-CTC TAC AGC TAT ATT GCC AGC CAC −3ʹ; CDK6-F: 5ʹ-TGG AGA CCT TCG AGC ACC-3ʹ; CDK6-F: 5ʹ-CAC TCC AGG CTC TGG AAC-3ʹ.
Western Blot Assay
The total protein was extracted and quantified by BCA assay (Beyotime, Nantong, China). Then, the lysate was analyzed by 12% SDS-PAGE, followed by transferring onto polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA, USA). After being blocked with 5% (w/v) non-fat dry milk, the membranes were incubated with antibodies, including anti-β-actin (1:2000), anti-E-cadherin (1:1000), anti-Vimentin (1:1000), and anti-α-SMA (1:1000) overnight at 4°C, respectively. All the antibodies were purchased from Abcam (Cambridge, UK). Then, the appropriate HRP-conjugated secondary antibodies (1:5000, Proteintech, Wuhan, China) were applied for 1 hour at RT. The protein bands were detected by ECL luminescence kit (Pierce, Rockford, IL, USA). Quantitative analysis was carried out on ImageJ 1.8.
Immunofluorescence Analysis
The cells were inoculated into a petri dish with pre-treated glass slides. When the density reached 80%, slides of cells were fixed with 4% paraformaldehyde and permeated with 0.5% Triton. Subsequently, the cells were washed with PBS and blocked with 3% (w/v) BSA at 4°C overnight. Then, the cell slides were incubated with primary antibodies against α-SMA (1:500 dilution) at 4°C overnight. Antibodies were purchased from Abcam (Shanghai, China). After incubating with FITC-conjugated secondary antibodies (1:200 dilution, Proteintech, Wuhan, China) at RT for 1 hour, the slides were observed with a fluorescence microscope (IX73, Olympus, Japan).
Cell Viability Assay
Cells were cultured in corresponding complete medium in 96-well plates. When the density reached 70%, cells were transfected as mention above. After 48 hours of transfection, 10 μL CCK-8 solution (Beyotime, Nantong, China) was added to each well. After incubating for 60 minutes, the absorbance was detected by a microplate reader at 450nm wavelength (Thermo Fisher, Massachusetts, USA).
Colony Formation Assay
The transfected SiHa and Hela cells were seeded into a 6-well plate (1×103 cells/well) at 37°C and cultured with 5% CO2. After two weeks, the cells were washed with phosphate buffer, fixed overnight with paraformaldehyde, and was stained with 0.4% crystal violet (Solarbio Science and Technology Ltd., Beijing, China) for 30 minutes. The colony counts were carried out under the inverted microscope.
Wound Healing Migration Assay
The transfected SiHa and Hela cells (3×106 cells/well) were seeded in a 6-well plate and grown to nearly 90% confluence. Subsequently, a linear scratch was made with a 200 μL pipette tip. After being washed with PBS three times, the cells were incubated with serum-free DMEM medium. All images were photographed at 0 and 24 h at 100×magnification and the size of the wound gap was measured. Each experiment was performed at least three times independently.
Transwell Chamber Assay
For the transwell migration and invasion assay, a 24-well transwell chamber that the insert membranes were coated with or without diluted Matrigel was used. The transfected SiHa and Hela cells (1×105 cells) were added to the upper chamber and cultured for 24 hours. Subsequently, the insert membranes were cut and stained with crystal violet (Solarbio Science and Technology Ltd., Beijing, China). The images were taken by an inverted microscope, and the number of invading cells was counted in three wells per group.
RNA Immunoprecipitation Assay
SiHa and Hela Cells (1×107) were added to 1mL IP buffer (50mM pH 7.4 Tris-HCl, 100mM NaCl, 0.5% NP-40, 0.5% sodium deoxycholate, 4% Protease Inhibitor Cocktail, 40 U/mL RNase inhibitor, and 20 μM MG132) resuspension cells and lysate on ice for 20 minutes. Then, the supernatant was ×10000g centrifuged at 4°C for 10 minutes, and the same amount of 2× SDS sample buffer was added as the input protein sample, or 1mL TRIzol was added as the input RNA sample. Each sample was incubated with 20μL protein A/G magnetic beads at 4°C for 1 hour. The beads were washed with IP buffer and then were added to the SDS sample buffer as the IP protein sample or were added to TRIzol as the IP RNA sample. The enrichment efficiency and the LINC00313 and miR-4677-3p level of binding RNA were detected by qPCR.
Luciferase Reporter Gene Assay
The wild-type (WT) LINC00313 transcript containing miR-4677-3p binding site and mutant (Mut) LINC00313 transcript with the mutated miR-4677-3p binding site were cloned into the psiCHECK-2 vector to construct psiCHECK-2-LINC00313-WT and psiCHECK-2-LINC00313-Mut reporter plasmid. Moreover, the 3ʹ-UTRs (wild type) and the corresponding mutant 3ʹ-UTRs of CDK6 were amplified by PCR. Obtained target fragments were cloned into the psiCHECK-2 reporter vector to construct psiCHECK-2-CDK6-WT and psiCHECK-2-CDK6-Mut reporter plasmid. SiHa cells were inoculated into 24-well plates at the concentration of 1 × 105 cells/cm2. When the cell density reached 70–80%, reporter gene plasmids, LINC00313 overexpression plasmids, and miR-4677-3p mimics or mimic control were co-transfected into SiHa cells by Lipofectamine 3000 (ThermoFisher, Shanghai, China). The cells were collected 48 hours after transfection, and luciferase activity was detected.
Statistical Analysis
Data were presented with mean ± SD. Pearson’s Correlation Analysis analyzed the correlation between the two groups. Student’s t-tests and one-way ANOVA with a Tukey post hoc test were used for statistical analysis by statistical software SPSS17.0. P < 0.05 was considered to be statistically significant.
Results
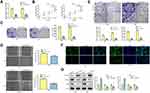
LINC00313 is Overexpressed in Cervical Carcinoma Tissues and Cells
In order to analyze the physiological expression, the levels of LINC00313 in 50 matched cervical carcinoma tissues and corresponding paracancerous tissues were detected by qRT-PCR. Then, the level of LINC00313 in tumor tissues was higher than that in paracancerous tissues (Figure 1A). Besides, in these 50 cases, the expression levels of LINC00313 in advanced cases (stage III and IV) was higher than that in early-stage cases (stage I and II) (P < 0.01; Figure 1B). Further data revealed that overall survival rates of cervical cancer patients with high LINC00313 levels were decreased, compared to the low LINC00313 levels group (Figure 1C). To further determine whether there had a correlation between LINC00313 Expression and patients, the pathology parameters were collected (Table 1). We found that LINC00313 expression was not significantly correlated with patient age, and tissue histological subtype (p>0.05), but it showed close correlation with FIGO stage, or tumour grading (p<0.01). At the cellular levels, compared with H8 cells (Human cervical epithelial cell line), the levels of LINC00313 in CaSki, SiHa, Hela, and C33A cells were significantly up-regulated (P < 0.01; Figure 1D), indicating that up-regulation of LINC00313 is positively correlated with the cervical carcinoma progression. Our data suggest that the high expression of LINC00313 has adverse effects on the prognosis of cervical carcinoma.
 |
Table 1 Clinical and Pathological Parameters for Age, FIGO-Classification, Grading and Histological Subtype |
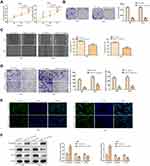
Knockdown of LINC00313 Inhibits the Progression of Cervical Carcinoma Cells
Next, we addressed the regulatory effects of LINC00313 on the aggressive phenotype of cervical carcinoma. To this end, we knocked down the expression of LINC00313 in SiHa and Hela cells. The transfection efficiency was confirmed by qRT-PCR (Figure 2A). Functionally, CCK8 assay and colony formation assay revealed that the proliferation of SiHa and Hela cells was inhibited after the knockdown of LINC00313 (Figure 2B and C). As revealed in Figure 2D, the wound closure percentages were decreased after the knockdown of LINC00313, compared with that of the control group (P < 0.01). Moreover, results showed that the knockdown of LINC00313 weakened the migration of SiHa and Hela cells (Figure 2E). The fluorescence intensity of α-SMA was decreased after the knockdown of LINC00313 (Figure 2F). Furthermore, we evaluated the expression levels of EMT hallmarks by Western Blot, including E-cadherin, Vimentin, and α-SMA. After the knockdown of LINC00313, the protein expression of E-cadherin was increased, while the protein expression of Vimentin and α-SMA were decreased (Figure 2G). These results indicated that abnormal over-expression of LINC00313 promotes the progression of cervical carcinoma cells.
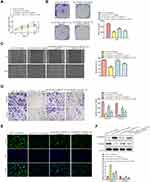
LINC00313 Serves as a Sponge of miR-4677-3p
To further explore the mechanism of LINC00313 involved in cervical carcinoma, RNA was isolated from the nucleus and cytoplasm to detect the expression of LINC00313 in the nucleus/cytoplasm. It was found that LINC00313 was more localized in the cytoplasm in both SiHa and Hela cells (Figure 3A). Moreover, we predicted that miR-4677-3p was the target of LINC00313 using the biological target prediction online tool TargetScan (http://www.targetscan.org/vert_72/), miRcode (http://www.mircode.org/), and Lncrnadb (http://lncrnadb.org/) (Figure 3B). The transfection efficiency of miR-4677-3p mimics was measured (Figure 3C). Then, Luciferase reporter assay showed that miR-4677-3p decreased the luciferase activity of LINC00313, while this inhibition was blocked by mutation of the potential binding domains (Figure 3D). Furthermore, the RIP assay showed that LINC00313 and miR-4677-3p were enriched preferentially in miRNPs containing AGO2 compared with anti-IgG immunoprecipitates (Figure 3E). We further analyzed the expression level of miR-4677-3p in tumor tissues and healthy tissues. As a result, the level of miR-4677-3p in tumor tissues was lower than that in paracancerous tissues (Figure 3F). Consistent with this, the expression levels of miR-4677-3p in CaSki, SiHa, Hela, and C33A cells were lower than those in H8 cells (Figure 3G). Furthermore, the expression levels of miR-4677-3p were significantly up-regulated in response to the transfection of sh-LINC00313 (Figure 3H). As shown in Table 1, we found that miR-4677-3p expression was not significantly correlated with patient age, and tissue histological subtype (p>0.05), but it showed close correlation with FIGO stage, or tumour grading (p<0.01). Meanwhile, Pearson’s Correlation Analysis showed that the correlation between LINC00313 and miR-4677-3p was negatively significant (P = 0.0498, Figure 3I). Overall, these results suggest that LINC00313 serves as a sponge of miR-4677-3p in cervical cancer cells.
Overexpression of miR-4677-3p Inhibits the Progression, Migration, and EMT of Cervical Carcinoma Cells
Next, we further explored the mechanism of miR-4677-3p involved in cervical cancer. Functionally, CCK8 and colony formation assays suggested that the up-regulation of miR-4677-3p inhibited the progression of SiHa and Hela cells (Figure 4A and B). Furthermore, the wound closure percentages were decreased in these cells with overexpression of miR-4677-3p, compared with that of the control group (Figure 4C). Moreover, the results showed that the up-regulation of miR-4677-3p weakened the migration of SiHa and Hela cells (Figure 4D). The fluorescence intensity of α-SMA was expectedly decreased in these cells with miR-4677-3p mimics transfection (Figure 4E). Furthermore, we evaluated the expression levels of EMT hallmarks by Western Blot. After the transfection of miR-4677-3p mimics, the protein expression of E-cadherin was increased, while the protein expression of Vimentin and α-SMA were decreased in SiHa and Hela cells (Figure 4F). These results indicated that abnormal fluctuation of the LINC00313/miR-4677-3p regulatory axis leads to the accelerated progression of cervical carcinoma cells.
CDK6 is the Target of miR-4677-3p
Because lncRNAs and miRNAs are generally not direct effector molecules of biological process, the downstream protein of the LINC00313/miR-4677-3p axis was further explored. Then, we predicted that CDK6 was the target of miR-4677-3p using the online tool TargetScan ((http://www.targetscan.org/vert_72/) and miRcode (http://www.mircode.org/) (Figure 5A). Given that CDK6 is a critical cell cycle regulator for the excessive proliferation of cancer cells,13 we further confirmed this prediction. The luciferase reporter assay revealed that miR-4677-3p decreased the luciferase activity of CDK6, while this inhibition was blocked by mutation of the potential binding domains (Figure 5B). Moreover, the mRNA levels and protein expression of CDK6 were significantly down-regulated after the transfection of miR-4677-3p mimics (Figure 5C and D). Besides, we analyzed the CDK6 expression in tumor tissues and healthy tissues. The results revealed that the level of CDK6 in tumor tissues was higher than that in paracancerous tissues (P < 0.01, Figure 5E). Consistent with this trend, the expression of CDK6 in CaSki, SiHa, Hela, and C33A cells was lower than those in H8 cells (Figure 5F). The above data suggest that miR-4677-3p acts as a sponge of CDK6 in cervical carcinoma. TAs shown in Table 1, we found that CDK6 expression was not significantly correlated with patient age, and tissue histological subtype (p>0.05), but it showed close correlation with FIGO stage, or tumour grading (p<0.01). Meanwhile, Pearson’s Correlation Analysis showed that the correlation between CDK6 and miR-4677-3p was negatively significant (P = 0.0004, Figure 5G). Of note, the correlation between LINC00313 and CDK6 was also positively significant (P = 0.0022, Figure 5H). Overall, these results demonstrated that LINC00313 up-regulated the expression of CDK6 through the down-regulation of miR-4677-3p.
LINC00313 Accelerates the Progression of Cervical Carcinoma Through miR-4677-3p/CDK6 Regulatory Axis
To further confirm the regulatory effects of the LINC00313/miR-4677-3p/CDK6 axis, we constructed LINC00313-silenced cervical cancer cells. CCK8 assay revealed that the LINC00313 knockdown inhibited the proliferation of SiHa and Hela cells (P < 0.01; Figure 6A), whereas miR-4677-3p inhibitors significantly neutralized LINC00313 silencing-inhibited cell proliferation (P < 0.01). Moreover, the cell viability of sh-LINC00313/miR-4677-3p inhibitors/sh-CDK6 group was higher than that of the sh-LINC00313/miR-4677-3p inhibitors group (P < 0.01). The trend of subsequent colony formation assay was consistent with these results (Figure 6B). Furthermore, we measured the regulatory effects of the miR-4677-3p/CDK6 axis on the migration of LINC00313-silenced cervical carcinoma cells. As demonstrated in Figure 6C and D, miR-4677-3p inhibition promoted the metastasis potential of LINC00313-silenced SiHa and Hela cells (P < 0.01), whereas CDK6 knockdown significantly reversed this trend (P < 0.01). At the molecular level, miR-4677-3p inhibition reversed the change of α-SMA fluorescence intensity caused by LINC00313 silencing, while CDK6 knockdown partially blocked the miR-4677-3p inhibition (Figure 6E). As expected, the trend of following EMT hallmarks expression was consistent with these results (Figure 6F). The above data suggested that the miR-4677-3p/CDK6 axis is downstream of LINC00313, indicating that the regulatory axis of LINC00313/miR-4677-3p/CDK6 materially existed in cervical cancer cells.
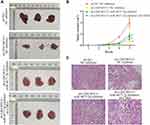
LINC00313 Promotes Cervical Cell Growth in vivo
To further study the effects of LINC00313 on the tumorigenicity of cervical mice models, the transfected-Hela cells were subcutaneously injected into the right side of Balb/c nude mice. After five weeks, the tumor volume was observed. As revealed in Figure 7A and B, the tumors of the sh-LINC00313 + NC inhibitor group were smaller than that of the control group (P < 0.01), whereas the sh-LINC00313 + miR-4677-3p inhibitor group were observed the opposite trend (P < 0.05). As shown in H&E staining of tumor tissues, the density of tumor cells of the sh-LINC00313+NC inhibitor group was lower, compared to the control group (Figure 7C). Moreover, the density of tumor cells of the sh-LINC00313 + miR-4677-3p inhibitor group was higher, compared to the sh-LINC00313 + NC inhibitor group. These results suggested that LINC00313 promotes cervical cell proliferation in vivo.
Discussion
To date, considerable progress has been made in the prevention, diagnosis, and treatment of cervical cancer. However, due to the metastasis and recurrence of cervical cancer, the effects of traditional surgery, radiotherapy, and chemotherapy are still limited in the treatment of patients with advanced metastatic cervical cancer.14 If patients with cervical cancer can get early diagnosis and early treatment, the 5-year survival rate can be as high as 90%.15,16 However, the early symptoms of cervical cancer are not obvious. 70% of the patients with cervical cancer are in the middle and late stage of the disease, about 31% of the cervical cancer will relapse after treatment, and the vast majority of recurrence occurs within 2 years after the diagnosis of the disease.17 At the same time, the treatment and prognosis of recurrent cervical cancer is very poor, and the 1-year survival rate is only 8% 12%.18 Therefore, it is urgent to find effective therapeutic targets and prognostic biomarkers for the diagnosis and treatment of cervical cancer.
Researches on lncRNA has received more and more attention and has become the hotspot of tumor research.19,20 The imbalance of lncRNA expression has been widely reported in cervical carcinoma, and lncRNA plays a crucial role in tumorigenesis, invasion, metastasis, and recurrence.21 For instance, the expression of circulating lncRNA-HOTAIR is positively regulated in cervical carcinoma and is increased significantly with the development of tumor stage, lymph node metastasis, and deep myometrial invasion.22,23 Because the up-regulation of lncRNA-HOTAIR is related to tumor recurrence and survival rate reduction, and it has been recognized as a prognostic marker of cervical cancer.24 The up-regulation of another lncRNA, lncRNA-H19, also has been found in cervical cancer patients.25 In this study, our findings revealed that LINC00313 was up-regulated in cervical carcinoma tissues and cells. Functionally, knockdown of LINC00313 arrested the progression, migration, and EMT of cervical carcinoma in vitro. These findings indicated that the abnormal expression of LINC00313 is related to the progression of cervical cancer.
MiRNAs are a class of non-coding single-stranded RNAs molecules encoded by endogenous genes with a length of about 22 nucleotides. MiRNAs can reduce protein expression by blocking the translation of mRNAs and promoting the degradation of mRNAs. Current studies have shown that miRNAs can directly regulate more than 60% of the protein-coding genes.26 As a negative regulator of gene regulation, miRNAs are involved in the development and treatment of various cancers.27 In this study, the luciferase report assays showed that LINC00313 sponged the miR-4677-3p. Further results showed that the expression of miR-4677-3p was up-regulated in sh-LINC00313 transfected cells. Compared with healthy tissues cells, the expression of miR-4677-3p was down-regulated in cervical cancer tissues and cells. In a recent study, Zhong et al,28 found that lncRNA TTN-AS1 promoted the migration of lung adenocarcinoma by inhibiting the expression of miR-4677-3p. Consistent with this finding, our findings also suggested that LINC00313 inhibits the miR-4677-3p expression and promotes cervical carcinoma progression.
Cyclin-dependent kinase 6 (CDK6) is a critical cell cycle regulator for the excessive proliferation of tumor cells, which forms a complex with cyclin D1 necessary for G1/S phase transformation.13 Given the excellent outcomes in preclinical and clinical,29–31 CDK6 inhibitors have become a promising therapeutic strategy for cancer patients with CDK6 overexpression. Overexpression of CDK6 in cervical cancer patients was also observed,30,32 which is consistent with our results. In this study, we found that there was a significant correlation between the expression of LINC00313, miR-4677-3p, and CDK6, respectively. Moreover, the luciferase report assays showed that miR-4677-3p inhibited the activity of CDK6 3ʹUTR. The mRNA and protein levels of CDK6 were down-regulated correspondingly to miR-4677-3p overexpression. Further rescue assay showed that miR-4677-3p inhibitors blocked the inhibitory effect of sh-LINC00313 on cervical cancer cells, while the knockdown of CDK6 could reverse this process. These findings suggested that the LINC00313/miR-4677-3p axis materially promotes the expression of CDK6, which leads to the progression of cervical cancer cells.
In conclusion, this study revealed that LINC00313 was overexpressed in cervical carcinoma tissues and cells. LINC00313 accelerated cervical carcinoma cell proliferation and metastasis by targeting the miR-4677-3p/CDK6 axis. Moreover, knockdown of LINC00313 inhibited the proliferation and metastasis potential in vitro, which was reversed by inhibition of the miR-4677-3p/CDK6 regulatory axis. Our findings demonstrated that LINC00313 accelerated the progression of cervical carcinoma via regulation of the miR-4677-3p/CDK6 axis.
Ethical Approval
This study was carried out based on the Ethics Committee of Nanjing Maternity and Child Health Care Hospital and the clinical analysis were performed according to the principles of the Helsinki Declaration. Written informed consent was obtained from all patients.
Funding
This work was supported by Nanjing Medical Science and Technique Development Foundation (No. YKK18157).
Disclosure
The authors declare that there are no conflicts of interest.
References
1. Vaccarella S, Laversanne M, Ferlay J, et al. Cervical cancer in Africa, Latin America and the Caribbean and Asia: regional inequalities and changing trends. Int J Cancer. 2017;141:1997–2001. doi:10.1002/ijc.30901
2. Ginsburg O, Bray F, Coleman MP, et al. The global burden of women’s cancers: a grand challenge in global health. Lancet. 2017;389:847–860. doi:10.1016/S0140-6736(16)31392-7
3. McGrath MJ, Binge LC, Sriratana A, et al. Regulation of the transcriptional coactivator FHL2 licenses activation of the androgen receptor in castrate-resistant prostate cancer. Cancer Res. 2013;73:5066–5079. doi:10.1158/0008-5472.CAN-12-4520
4. Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi:10.1016/j.tibs.2013.10.002
5. Wu F, Sui Y, Wang Y, et al. Long noncoding RNA SNHG7, a molecular sponge for microRNA-485, promotes the aggressive behavior of cervical cancer by regulating PAK4. Onco Targets Ther. 2020;13:685–699. doi:10.2147/OTT.S232542
6. Shen X, Zhao W, Zhang Y, et al. Long non-coding RNA-NEAT1 promotes cell migration and invasion via regulating miR-124/NF-κB pathway in cervical cancer. Onco Targets Ther. 2020;13:3265–3276. doi:10.2147/OTT.S220306
7. Chen L, Zhang X, Han B, et al. Long noncoding RNA SNHG12 indicates the prognosis and accelerates tumorigenesis of diffuse large B-Cell lymphoma through sponging microR-195. Onco Targets Ther. 2020;13:5563–5574. doi:10.2147/OTT.S249429
8. Shen F, Zheng H, Zhou L, et al. Overexpression of MALAT1 contributes to cervical cancer progression by acting as a sponge of miR-429. J Cell Physiol. 2019;234:11219–11226. doi:10.1002/jcp.27772
9. Wang N, Hou MS, Zhan Y, et al. MALAT1 promotes cisplatin resistance in cervical cancer by activating the PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:7653–7659. doi:10.26355/eurrev_201811_16382
10. Gao J, Liu L, Li G, et al. LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int J Biol Macromol. 2019;126:994–1001. doi:10.1016/j.ijbiomac.2018.12.176
11. Yao T, Lu R, Zhang J, et al. Growth arrest‐specific 5 attenuates cisplatin‐induced apoptosis in cervical cancer by regulating STAT3 signaling via miR‐21. J Cell Physiol. 2018;234:9605–9615. doi:10.1002/jcp.27647
12. Song C, Fan B, Xiao Z. Overexpression of ALK4 inhibits cell proliferation and migration through the inactivation of JAK/STAT3 signaling pathway in glioma. Biomed Pharmacother. 2018;98:440–445. doi:10.1016/j.biopha.2017.12.039
13. Handschick K, Beuerlein K, Jurida L, et al. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-κB-dependent gene expression. Mol Cell. 2014;53:193–208. doi:10.1016/j.molcel.2013.12.002
14. Refaei M, Dehghan NN, Khakbazan Z, et al. Exploring effective contextual factors for regular cervical cancer screening in Iranian women: a qualitative study. Asian Pac J Cancer Prev. 2018;19:533–539. doi:10.22034/APJCP.2018.19.2.533
15. The LO. Global elimination of cervical cancer is achievable-with commitment. Lancet Oncol. 2019;20:1467. doi:10.1016/S1470-2045(19)30647-3
16. Zhao F, Qiao Y. Cervical cancer prevention in China: a key to cancer control. Lancet. 2019;393:969–970. doi:10.1016/S0140-6736(18)32849-6
17. Brisson M, Kim JJ, Canfell K, et al. Impact of HPV vaccination and cervical screening on cervical cancer elimination: a comparative modelling analysis in 78 low-income and lower-middle-income countries. Lancet. 2020;395:575–590. doi:10.1016/S0140-6736(20)30068-4
18. Cohen PA, Jhingran A, Oaknin A, et al. Cervical cancer. Lancet. 2019;393:169–182. doi:10.1016/S0140-6736(18)32470-X
19. Bogani G, Leone Roberti Maggiore U, Signorelli M, et al. The role of human papillomavirus vaccines in cervical cancer: prevention and treatment. Crit Rev Oncol Hematol. 2018;122:92–97. doi:10.1016/j.critrevonc.2017.12.017
20. Dong J, Su M, Chang W, et al. Long non-coding RNAs on the stage of cervical cancer. Oncol Rep. 2017;38:1923–1931. doi:10.3892/or.2017.5905
21. Li TF, Liu J, Fu SJ. The interaction of long non-coding RNA MIAT and miR-133 play a role in the proliferation and metastasis of pancreatic carcinoma. Biomed Pharmacother. 2018;104:145–150. doi:10.1016/j.biopha.2018.05.043
22. Zheng P, Yin Z, Wu Y, et al. LncRNA HOTAIR promotes cell migration and invasion by regulating MKL1 via inhibition miR206 expression in HeLa cells. Cell Commun Signal. 2018;16:5. doi:10.1186/s12964-018-0216-3
23. Sharma Saha S, Roy Chowdhury R, Mondal NR, et al. Identification of genetic variation in the lncRNA HOTAIR associated with HPV16-related cervical cancer pathogenesis. Cell Oncol. 2016;39:559–572. doi:10.1007/s13402-016-0298-0
24. Kim HJ, Lee DW, Yim GW, et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi:10.3892/ijo.2014.2758
25. Roychowdhury A, Samadder S, Das P, et al. Deregulation of H19 is associated with cervical carcinoma. Genomics. 2020;112:961–970. doi:10.1016/j.ygeno.2019.06.012
26. Rivera-Barahona A, Pérez B, Richard E, et al. Role of miRNAs in human disease and inborn errors of metabolism. J Inherit Metab Dis. 2017;40:471–480. doi:10.1007/s10545-017-0018-6
27. Chen Y, Gao D, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliver Rev. 2015;81:128–141. doi:10.1016/j.addr.2014.05.009
28. Zhong Y, Wang J, Lv W, et al. LncRNA TTN‐AS1 drives invasion and migration of lung adenocarcinoma cells via modulation of miR‐4677‐3p/ZEB1 axis. J Cell Biochem. 2019;120:17131–17141. doi:10.1002/jcb.28973
29. Adkins D, Ley J, Neupane P, et al. Palbociclib and cetuximab in platinum-resistant and in cetuximab-resistant human papillomavirus-unrelated head and neck cancer: a multicentre, multigroup, Phase 2 trial. Lancet Oncol. 2019;20:1295–1305. doi:10.1016/S1470-2045(19)30405-X
30. Xiong Y, Li T, Assani G, et al. Ribociclib, a selective cyclin D kinase 4/6 inhibitor, inhibits proliferation and induces apoptosis of human cervical cancer in vitro and in vivo. Biomed Pharmacother. 2019;112:108602. doi:10.1016/j.biopha.2019.108602
31. Dickler MN, Tolaney SM, Rugo HS, et al. MONARCH 1, A Phase II study of abemaciclib, a CDK4 and CDK6 inhibitor, as a single agent, in patients with refractory HR+/HER2− Metastatic breast cancer. Clin Cancer Res. 2017;23:5218–5224. doi:10.1158/1078-0432.CCR-17-0754
32. Liu Q, Liu S, Wang X, et al. LncRNA MAGI2-AS3 is involved in cervical squamous cell carcinoma development through CDK6 up-regulation. Infect Agents Cancer. 2019;14:37. doi:10.1186/s13027-019-0238-5
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.