Back to Journals » Cancer Management and Research » Volume 13
Long Non-Coding RNA JPX Contributes to Tumorigenesis by Regulating miR-5195-3p/VEGFA in Non-Small Cell Lung Cancer
Authors Li G, Li X, Yuan C, Zhou C, Li X, Li J, Guo B
Received 24 March 2020
Accepted for publication 12 January 2021
Published 12 February 2021 Volume 2021:13 Pages 1477—1489
DOI https://doi.org/10.2147/CMAR.S255317
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Beicheng Sun
Guanglian Li, Xinrui Li, Chao Yuan, Caifeng Zhou, Xinxin Li, Jinfang Li, Bin Guo
Department of Oncology, The People’s Hospital of Shouguang, Weifang, People’s Republic of China
Correspondence: Bin Guo
Department of Oncology, The People’s Hospital of Shouguang, No. 1223, Jiankang Street, Shouguang, Shandong, 262700, People’s Republic of China
Tel +86 536 5225033
Email [email protected]
Background: Lung cancer is the most frequently diagnosed cancer. Of all lung cancers, 80– 85% are verified as non-small-cell lung cancer (NSCLC). Just proximal to X-inactive specific transcript (JPX), functions as lncRNA, contributed to tumor progression and suggested a poor prognosis in NSCLC. However, the pathogenesis of JPX involved in NSCLC is still unclear.
Methods: The expressions of JPX, miR-5195-3p, and Vascular endothelial growth factor A (VEGFA) were detected by real-time quantitative polymerase chain reaction (RT-qPCR). Proliferation, colony number, apoptosis, invasion, and migration were analyzed by Cell Counting Kit-8 (CCK-8), colony formation, flow cytometry, transwell, and wound healing assays, severally. The protein levels of VEGFA, E-cadherin, N-cadherin, and Vimentin were detected by Western blot assay. The interaction between JPX, miR-5195-3p and VEGFA was predicted by starBase, and then verified by the dual-luciferase reporter, RNA Immunoprecipitation (RIP) and RNA pull-down assay. The biological role of JPX on NSCLC tumor growth was assessed by the xenograft tumor model in vivo.
Results: JPX and VEGFA were upregulated, and miR-5195-3p was downregulated in NSCLC. JPX induced proliferation, colony number, invasion, migration, epithelial–mesenchymal transition (EMT), and inhibited apoptosis of NSCLC cells. JPX is directly bound to miR-5195-3p. JPX regulated NSCLC cell proliferation, apoptosis and EMT by modulating miR-5195-3p. miR-5195-3p hindered NSCLC cells proliferation, EMT and accelerated apoptosis by directly targeting VEGFA. JPX silencing hindered the cell growth of NSCLC in vivo.
Conclusion: JPX facilitated proliferation, colony number, invasion, migration, EMT, and repressed apoptosis by miR-5195-3p/VEGFA axis, offering a possible therapeutic strategy for NSCLC.
Keywords: lncRNA JPX, miR-5195-3p, VEGFA, lung cancer, proliferation, apoptosis, EMT
Introduction
Lung cancer, an incurable malignancy, is the leading cause of cancer death in adults worldwide. According to statistics in 2018, about 2.1 million new lung cancer cases and 1.8 million deaths occurred in the world.1 Noteworthy, 80–85% of all lung cancers were identified as non-small-cell lung cancer (NSCLC).2 Despite the substantial progress in detection and therapeutic approaches, the 5-year survival rate of lung cancer patients was less than 15%.3 Undoubtedly, the exploration of the molecular mechanism underlying NSCLC progression for identifying potential novel therapeutic targets is highly desirable.
Long non-coding RNA (lncRNA) is identified as a vital subgroup of non-coding RNA, which is longer than 200 nucleotides in length with no or a limited protein-coding capacity.4 Accumulating evidence indicated lncRNA is closely related to a serial of biological and cellular processes, such as gene expression, protein activity, tumorigenesis, and metastasis.5,6 Previous reporters have shown that lncRNAs could exert the function as oncogenes or tumor suppressions in a variety of human cancers,7 including NSCLC. For example, Dong et al confirmed that knockdown of lncRNA Gm15290 inhibited NSCLC cell development by binding to miR-615-5p.8 Miao et al reported that lncRNA HAND2-AS1 could act as an inhibitory effect by suppressing cell migration and invasion in NSCLC progression.9 Just proximal to X-inactive specific transcript (JPX), functions as lncRNA, has been proposed to activate (X-inactive specific transcript) XIST expression and is a crucial molecular switch in the inactivation of X-chromosome.10,11 Furthermore, a prior reporter has suggested that the overexpression of JPX contributed to tumor progression in NSCLC.12 However, the pathogenesis of JPX involved in NSCLC has not yet been fully elucidated.
MicroRNAs (miRNAs), short non-coding RNAs that consist of about 22 nucleotides, are implicated in the regulation of gene expression through mRNA degradation and gene translation repression.13 Accumulating reporters disclosed that miRNAs are extensively involved in multiple processes of diverse tumors, including NSCLC.14–16 Some studies displayed that miR-5195-3p are closely concerned with the tumor development in ovarian cancer and bladder cancer.17,18 Moreover, recent research confirmed that miR-5195-3p could hinder proliferation, migration, and invasion of NSCLC cells, implying that miR-5195-3p acted as an inhibiting effect in NSCLC development.19 Nevertheless, the potential mechanism of miR-5195-3p in NSCLC requires further investigation.
Vascular endothelial growth factor A (VEGFA), an endothelial cell-specific mitogen, plays a crucial role in angiogenesis.20,21 Some studies showed that VEGFA was associated with tumor progression in colorectal cancer and breast cancer.22,23 Additionally, VEGFA was reported to act as a carcinogenic factor by regulating cell growth and invasion in NSCLC,24 indicating the fatal role of VEGFA in NSCLC pathogenesis.
In this paper, we verified the relationship between JPX, miR-5195-3p, and VEGFA and investigated whether the involvement of JPX in NSCLC function was mediated by miR-5195-3p/VEGFA axis.
Materials and Methods
Clinical Specimens and Cell Culture
Sample of the NSCLC tissues (n=45) and corresponding normal lung tissues (n=45) were obtained from NSCLC patients who underwent surgical resection at the People’s Hospital of Shouguang. And the clinicopathological features of these patients are presented in Table 1. This work got approved by the ethics committee of the People’s Hospital of Shouguang and written informed consent was signed by every participant.
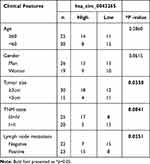 |
Table 1 Correlation Between lncRNA JPX Expression and Clinicopathological Parameters of NSCLC Patients (n = 45) |
Normal human lung cell line (BEAS-2B) was provided by Cell Bank of Chinese Academy of Sciences Committee on Type Culture Collection (Shanghai, China). NSCLC cell lines (NCI-H1299, A549, and NCI-H460) were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA). All NSCLC cells were incubated at 37°C with5% CO2 in a humidified air atmosphere and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% FBS (Gibco). BEAS-2B was cultured in Roswell Park Memorial Institute 1640 medium (RPMI 1640) (Invitrogen, Carlsbad, CA, USA) plus 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) at 37°C under 5% CO2 condition.
Cell Transfection
JPX siRNAs (si-JPX) and negative control si-RNA control (si-NC) were collected from RiboBio (Guangzhou, China). miR-5195-3p mimic (miR-5195-3p), miR-5195-3p inhibitor (anti-miR-5195-3p) and their corresponding negative controls (NC, anti-NC) were synthesized and purified by Genechem (Suzhou China). JPX or VEGFA overexpression vector was generated by inserting a full-length of JPX or VEGFA cDNA sequence into pcDNA3.1 empty vector (pcDNA, Invitrogen, Carlsbad, CA, USA), named as JPX-pcDNA3.1 (pcDNA-JPX), VEGFA-pcDNA3.1 (VEGFA). All oligonucleotides and plasmids were transfected into NCI-H1299 and A549 in line with the manufacturer’s protocol of Lipofectamine 2000 reagents (Invitrogen).
RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was isolated from NSCLC samples and cell lines referring to the producer’s instructions of TRIzol reagent (Invitrogen). Then, Moloney murine leukemia virus (M-MLV) reverse transcriptase (Takara, Shiga, Japan) was applied to synthesize cDNA. The RT-qPCR analysis was implemented on a StepOne Real-time PCR System (Applied Biosystems, Foster City, CA, USA) by using SYBR® PremixEx TaqTM kit (Takara) or the SYBRPrimeScriptTM miRNA RT-qPCR Kit (Takara). The 2–∆∆Ct method was executed to process the relative gene expression levels. House-keeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 small nuclear RNA (snRNA) acted as an internal control. The primer sequences in RT-qPCR were listed as follows:
JPX: 5ʹ-GCACCACCAGGCTTCTGTAAC-3ʹ (sense), 5ʹ-GGGCATGTTCATTAATTGGCCAG-3ʹ (antisense);
miR-5195-3p: 5ʹ-TAGCAGACTCTTATGATG −3ʹ (sense), 5ʹ-TGGTGGAGTCGTCGTG-3ʹ (antisense);
VEGFA: 5ʹ-TTTCTGCTGTCTTGGGTGCATTGG-3ʹ (sense), 5ʹ-ACCACTTCGTGATGATTCTGCCCT −3ʹ (antisense);
U6: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (sense), 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (antisense);
GAPDH: 5ʹ-GTGGACCTGACCTGCCGTCT-3ʹ (sense), 5ʹ-GGAGGAGTGGGTGTCGCTGT-3ʹ (antisense).
Cell Viability Assay
In this study, the Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology, Shanghai, China) was employed to assess cell viability in NSCLC cells in line with the supplier’s direction. After transfection, 3 × 103 cells/well NCI-H1299 and A549 cells were seeded into 96-well plates and cultured incubated for 0 h, 24 h, 48 h, or 72 h, severally. Next, these cells were treated with 10µL CCK-8 solution (Beyotime Institute of Biotechnology) for another 4 h. Finally, the optical density (OD) value was measured by using microplate reader.
Colony Formation Assay
In short, transfected NCI-H1299 and A549 cells (5×102 cells/well) in six-well plates were incubated for 2 weeks at 37°C. After discarding the medium, the cells were fixed with methyl alcohol for 15 min at −20°C, and then stained in 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at 37°C. Finally, a microscopy (Nikon, Tokyo, Japan) was used to acquire the image of the visible colonies per well, followed by calculation with Image J (NIH, Bethesda, MD, USA).
Cell Apoptosis Assay
The apoptosis rate of NCI-H1299 and A549 cells was detected by Annexin (V-fluorescein isothiocyanate) V-FITC/Propidium Iodide (PI) (Selleck, Shanghai, China) as per the user’s guidebook. Generally, transfected NCI-H1299 and A549 cells were pelleted and washed twice with pre-cold phosphate-buffered saline (PBS, Invitrogen). After resuspension in binding buffer, 5 μL Annexin V-FITC/PI (Selleck) was applied to the stain cells. At last, apoptosis NSCLC cells were measured with FACSan flow cytometry (BD Bioscience, San Jose, CA, USA) referring to the user’s guidebook.
Cell Invasion Assay
In this assay, the measurement of invasion was conducted using 24-well transwell chambers (Costar, Cambridge, Massachusetts, USA) coated with Matrigel (BD Bioscience, Franklin Lakes, NJ, USA). In general, transfected cells were resuspended in a serum-free medium at a density of 1×106 cells/well, followed by an introduction into the upper chamber. And the lower counterparts were filled up with the medium containing10% FBS (as a chemoattractant). Twenty-four hour later, the invading cells on the lower membrane surface were stained with 0.1%crystal violet (Sigma-Aldrich) for 15 min being fixed in 4% paraformaldehyde (Sigma-Aldrich) for 15 min. At last, a microscope (Nikon, magnification×100) was performed to photograph and count the stained cells.
Wound Healing Assay
The assessment of migratory rate in NCI-H1299 and A549 cells was carried out using a wound-healing assay. Briefly, transfected cells were plated into 6-well plates at 90–95% confluency. And then, wounds were scratched with a sterile pipette tip (record 0 h), followed by washing twice with PBS. After addition to the serum-free culture medium for 24 h, the analysis of the wound closing procedure was conducted with a light microscope (Nikon) and Image J (NIH).
Western Blot Assay
In brief, NCI-H1299 and A549 cells were collected and then lysed by using RIPA buffer (Beyotime, Shanghai, China) for obtaining total protein. Afterward, denatured proteins were incubated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 10%), transferred onto polyvinylidene fluoride (PVDF) membranes (Amer-sham Pharmacia, Piscataway, NJ, USA), and incubated with primary antibody against VEGFA (1:200, ab31745, Abcam, Cambridge, UK), E-cadherin (1:1000, sc-7870, Santa Cruz Biotechnology, Santa Cruz, CA, USA), N-cadherin (1:200, sc-7939, Santa Cruz Biotechnology), Vimentin (1:500, sc-5565, Santa Cruz Biotechnology) and GAPDH (1:5000, ab8245, Abcam) all-night at 4°C. The next day, the proteins were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Abcam) and detected by enhanced chemiluminescence (Applygen Technologies, Beijing, China).
Dual-Luciferase Reporter Assay
Partial sequences of JPX and VEGFA 3ʹUTR possessing wild-type or mutant-type putative miR-5195-3p targeting sites were amplified and cloned into luciferase reporter pmirGLO vector (Promega, Madison, Wisconsin, USA), termed JPX-wt/mut, VEGFA-wt/mut 3ʹUTR reporter plasmids. Subsequently, NCI-H1299 and A549 cells co-transfected with the constructed reporter plasmids and miR-5195-3p or anti-miR-5195-3p by Lipofectamine 2000 reagents (Invitrogen). Analysis of their luciferase activities was performed by using a dual-luciferase reporter assay kit (Beckman Coulter, Fullerton, CA, USA) at 48 h post-transfection.
RNA Immunoprecipitation (RIP) and RNA-IP Assays
To validate the interaction between miR-5195-3p and JPX or VEGFA, RIP assays were performed in accordance with the user’s guidebook of the Magna RIP RNA-binding protein Immunoprecipitation kit (Millipore, Billerica, MA, USA). NCI-H1299 and A549 cells were cultured for 48 h, and lysed with RIP lysis buffer embracing magnetic beads conjugated with anti-Argonaute2 (Ago2) antibody (1: 1000, ab32381, Abcam) or negative control IgG (Millipore) at 4°C for 6 h. After the digestion of proteinase K, immunoprecipitated RNA was segregated. RT-qPCR was used for analyzing the purified RNA. For RNA-IP assay, cells were lysed with RIP lysis buffer, followed by incubation with the beads conjugated with VEGFA antibody (1:1000, ab39250, Abcam) or IgG at 4°C overnight. And the following steps are consistent with the RIP assay.
RNA Pull-Down Assay
Generally, NCI-H1299 and A549 cells were transfected with biotinylated miR-5195-3p (bio-miR-5195-3p) or control NC (bio-NC) by Lipofectamine RNAiMax Reagent (Thermo Fisher Scientific). And then these transfected cells were harvested and sonicated. Remaining cell lysates were maintained with C-1 Magnetic Beads (Thermo Fisher Scientific) for 2 h at 4°C to pull down the biotin-coupled RNA complex. An RNeasy Mini Kit (Qiagen, Hilden, Germany) was used to purify these mixtures. At last, the RNA enrichment was detected using RT-qPCR.
In vivo Assay
Five-week-old male BALB/C nude mice were purchased from the Vital River Laboratory (Beijing, China) in this vivo experiment, which was approved by the Animal Ethics Committee of the People’s Hospital of Shouguang. Whereafter, a total of 5×106 NCI-H1299 cells stably transfected with LV-sh-NC or LV-sh-JPX (Genechem) were injected subcutaneously into the flank of the nude mice with 6 mice in each group, and the tumor volume every 5 days. A month later, all mice were sacrificed, and the resected tumor masses were harvested for weighting and further analysis.
Statistical Analysis
Every experiment was independently repeated three times. The values were presented as mean ± standard deviation (SD). Data were analyzed with SPSS 18.0 software (IBM, Chicago, IL, USA). Expression association of JPX, miR-5195-3p, and VEGFA was analyzed using Pearson correlation analysis. Student’s t-test or one way of variance (ANOVA) was implemented for analysis of significant difference (P< 0.05).
Results
JPX Induced Proliferation, Invasion, Migration, EMT, and Inhibited Apoptosis of NSCLC Cells in vitro
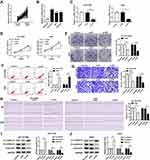
We first investigated the level of JPX in NSCLC tissues and normal tissues. As compared with normal tissues (n=45), JPX expression was notably elevated in NSCLC tumor tissues (n=45) (Figure 1A). Analyzing the clinical data of NSCLC patients, we found that the expression of JPX was correlated with the patient’s tumor size, TNM state, and lymph node metastasis (Table 1). Moreover, we further validated that JPX expression was remarkably increased in NSCLC cell lines (NCI-H1299, A549, NCI-H460) in contrast to normal human lung cell line (BEAS-2B) (Figure 1B). Then, to further explore the effect of JPX on NSCLC cells, we knocked down JPX in NCI-H1299 and A549 cells. As displayed in Figure 1C, JPX level was markedly downregulated in si-JPX-transfected NCI-H1299 and A549 cells versus si-NC-transfected cells. Thus, we further explored the biological function of JPX in NSCLC progression by knocking down systems. The results of CCK-8 and cell colony formation showed that depletion of JPX strikingly repressed the proliferative ability of NCI-H1299 and A549 cells (Figure 1D and E). Moreover, flow cytometry assay revealed that cell apoptosis was apparently improved after downregulating the expression of JPX in NCI-H1299 and A549 cells (Figure 1F). Apart from that, the reduced invasion and migration were viewed due to the deficiency of JPX in NCI-H1299 and A549 cells (Figure 1G and H). Whereafter, we further probed that the role of JPX on epithelial–mesenchymal transition (EMT) of NCI-H1299 and A549 cells. The results presented that knockdown of JPX led to an evident enhancement in E-cadherin protein level, and an obvious reduction in N-cadherin and Vimentin protein levels, indicating JPX promoted EMT in NCI-H1299 and A549 cells (Figure 1I and J). On the contrary, our data indicated that the overexpression of JPX promoted cell proliferation ability, invasion, migration, and repressed apoptosis of NCI-H1299 and A549 cells (Figure S1). Overall, these data indicated that the dysregulation of JPX might be involved with the NSCLC progression.
MiR-5195-3p Was a Direct Target of JPX in NSCLC Cells
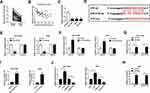
Accumulative evidence has suggested that lncRNAs serve as miRNA sponges to modulate gene expression post-transcriptional.25 Therefore, we investigated whether JPX regulated NSCLC development in a miRNA-mRNA dependent manner. First of all, we observed miR-5195-3p was lower expressed in NSCLC tissues and cells versus respective control groups (Figure 2A and C). Synchronously, miR-5195-3p level was inversely related to the level of JPX in NSCLC tissues (Figure 2B). Next, to ascertain whether JPX works as a molecular sponge of miR-5195-3p in NSCLC, bioinformatics analysis was carried out by starBase software. As illustrated in Figure 2D, miR-5195-3p harbored some complementary binding sites with JPX. To verify the interaction of miR-5195-3p with JPX was mediated by the putative binding site, a dual-luciferase reporter assay was performed in NSCLC. Data suggested that miR-5195-3p downregulation restrained the luciferase activity of JPX-wt reporter and miR-5195-3p overexpression reinforced the luciferase activity of JPX-wt reporter, however, overexpression and downregulation of miR-5195-3p had little effect on the luciferase activity of JPX-mut in NCI-H1299 and A549 cells (Figure 2E and F). Moreover, previous studies have suggested that lncRNA negatively regulated miRNA expression by combining with Ago2-containing RNA-induced silencing complex (RISC).26 To confirm the binding between JPX and miR-5195-3p, RIP assay was carried out in NCI-H1299 and A549 cells. Compared with anti-normal IgG group, cell extracts between JPX and miR-5195-3p were apparently enriched in anti-Ago2 group (Figure 2G and H), supporting the binding between JPX and miR-5195-3p. Besides, we conducted RNA pull-down assay to further determine the interaction between JPX and miR-5195-3p. Consistent with bioinformatics analysis, luciferase assay and RIP assay, we found that JPX enrichments were distinctly upregulated in miR-5195-3p group in comparison with NC group (Figure 2I), identifying the interaction existed between them. In addition, RT-qPCR results also demonstrated that the negative correlation between JPX and miR-5195-3p in NCI-H1299 and A549 cells (Figure 2J). Thus, JPX interacts with miR-5195-3p to repress its expression.
MiR-5195-3p Partially Reversed the Effect of JPX on Proliferation, Apoptosis and EMT of NSCLC Cells
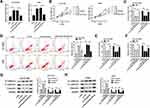
Next, to further probe the mechanism of JPX involved in NSCLC progression, NCI-H1299 and A549 cells were transfected with si-JPX and anti-miR-5195-3p. The transfection efficiency was measured and exhibited in Figure 3A. Functionally, data presented that cell proliferative ability impaired by knocking-down of JPX was partly recovered by the downregulation of miR-5195-3p in NCI-H1299 and A549 cells (Figure 3B and C). Then, flow cytometry analysis showed that silencing of miR-5195-3p partially abrogated the positive effect of JPX depletion on cell apoptosis of NCI-H1299 and A549 cells (Figure 3D). Meanwhile, si-JPX mediated declined in the abilities of invasion and migration was significantly mitigate by the deficiency of JPX in NCI-H1299 and A549 cells (Figure 3E and F). In addition, Western blot results pointed out that JPX downregulation accelerated E-cadherin protein level and reduced N-cadherin and Vimentin protein level, which was attenuated by suppression of miR-5195-3p (Figure 3G and H). Collectively, these data suggested that JPX contributed to NSCLC progression by targeting miR-5195-3p.
VEGFA as a Target of miR-5195-3p
Additionally, recent studies reported that VEGFA is highly expressed in lung cancer,27 which was also confirmed by our research. As presented in Figure 4A, VEGFA expression was upregulated in NSCLC tumor tissues in comparison to corresponding non-cancerous tissues. Similarly, we acknowledged that both mRNA level and protein level of VEGFA were increased in NCI-H1299 and A549 cells versus BEAS-2B cell (Figure 4C and D). Furthermore, the inverse relationship between miR-5195-3p and VEGFA was observed in NSCLC tissues (Figure 4B). Thus, we used online publicly available database starBase to predict the relationship between VEGFA and miR-5195-3p. As indicated in Figure 4E, VEGFA was found to have some binding sequences of miR-5195-3p. Then, luciferase activity analysis was used to validate the potential interaction. The luciferase reporter vectors both VEGFA-wt 3ʹUTR and VEGFA-mut 3ʹUTR were co-transfected with NC or miR-5195-3p into NCI-H1299 and A549 cells. Subsequent luciferase assays showed that ectopic expression of miR-5195-3p reduced the luciferase activity of VEGFA-wt, but had no obvious effect on the luciferase activity of VEGFA-mut (Figure 4F). Synchronously, to further confirm the binding between miR-5195-3p and VEGFA, we implemented RIP assays to pull down miRNA concerned with VEGFA by antibodies against Ago 2 in NCI-H1299 and A549 cells. As a result, miR-5195-3p and VEGFA were strikingly enriched in Ago 2 pellets when compared with the IgG control group (Figure 4G). Also, RNA-IP assay was performed to prove the interaction among VEGFA and miR-5195-3p or JPX. As displayed in Figure 4H, the enrichment of miR-5195-3p and JPX was improved in the anti-VEGFA groups relative to the IgG groups in NCI-H1299 and A549 cells. In a word, VEGFA could directly bind to miR-5195-3p and JPX in NSCLC cells. What’s more, we found that VEGFA expression level was positively correlated with JPX in NSCLC tissues (Figure 4I). Meanwhile, our data suggested that the overexpression of JPX could boost VEGFA protein level, inversely, the knockdown of JPX could dampen VEGFA level in NCI-H1299 and A549 cells (Figure 4J). Importantly, Western blot results revealed that reintroduction of JPX effectively counteracted miR-5195-3p overexpression-caused decline in VEGFA protein level in NCI-H1299 and A549 cells (Figure 4K), suggesting that JPX could positively regulate VEGFA by interacting with miR-5195-3p.
MiR-5195-3p Suppressed Proliferation, EMT and Facilitated Apoptosis by Targeting VEGFA in NSCLC Cells
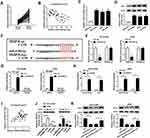
As mentioned above, we speculated that the miR-5195-3p/VEGFA axis may be implicated in the development of NSCLC. In order to validate the assumption, rescue assay was conducted in NSCLC cells. As exhibited in Figure 5A, upregulation of miR-5195-3p apparently reduced VEGFA expression, which was effectively mitigated by the co-transfection with pcDNA-VEGFA in NCI-H1299 and A549 cells. Functionally, CCK-8 results proved that the inhibition of cell proliferative ability induced by miR-5195-3p overexpression was weakened by upregulation of VEGFA in NCI-H1299 and A549 cells (Figure 5B and C). Then, flow cytometry analysis pointed out that the promotion of cell apoptosis caused by the high expression of miR-5195-3p was reversed by upregulating of VEGFA in NCI-H1299 and A549 cells (Figure 5D). Synchronously, the results from transwell and wound healing indicated that the forced expression of VEGFA could attenuate the suppression effect of miR-5195-3p on invasion and migration in NCI-H1299 and A549 cells (Figure 5E and F). Besides, Western blot assay exhibited that co-transfection of VEGFA prominently abrogated miR-5195-3p-triggered increase in E-cadherin protein level, and reduction in N-cadherin and Vimentin protein level (Figure 5G and H), testifying that miR-5195-3p blocked NSCLC cells EMT by regulating VEGFA partially overturned the suppressive effect of miR-5195-3p overexpression in NSCLC cells. All in all, miR-5195-3p could hinder NSCLC progression via modulating VEGFA.
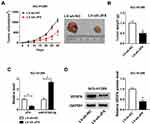
JPX Knockdown Suppressed NSCLC Tumor Growth in vivo
Additionally, to further explore the influence of JPX deficiency on tumor growth in vivo, we established mice xenograft models of NSCLC. As presented in Figure 6A and B, the tumor volume and weight were decreased in the presence of JPX knockdown, implying that the stable downregulation of JPX significantly blocked cell growth of NSCLC in vivo. Apart from that, our data indicated that the expression of JPX and VEGFA was declined, whereas miR-5195-3p level was enhanced in tumor tissues derived from the LV-sh-JPX group relative to the LV-sh-NC group (Figure 6C and D). That was to say, JPX deficiency could inhibit tumor growth in NSCLC in vivo.
Discussion
Recently, some studies indicate that lncRNAs perform as essential regulators of tumorigenesis in NSCLC, providing potential biomarkers for prognosis and therapy of NSCLC.28–30 JPX, located on the X chromosome, has been documented as an activator of the XIST and a molecular switch for X chromosome inactivation.10,11 In the recent several years, previous studies identified that JPX was highly expressed in NSCLC, and involved in tumor progression and indicated a poor prognosis.12 However, the biological function and molecular mechanisms of JPX are still not fully established.
Thus, in our study, we confirmed that JPX level was upregulated in NSCLC tissues and NSCLC cells relative to respective control groups. Subsequently, we further explored proliferation, apoptosis, and EMT in NSCLC cells transfected with si-JPX. The results suggested that JPX promoted proliferation, EMT and inhibited apoptosis of NSCLC cells. That is to say, JPX exerted a tumor-promoting role in the development of NSCLC.
MiR-5195-3p was reported to be downregulated, and exert a suppressive role in NSCLC.19 In this study, miR-5195-3p was viewed to be an obvious low expression in NSCLC tissues and cells, and negatively associated with JPX in NSCLC tissues. Hence, we further investigated whether JPX could use function in NSCLC by regulating miR-5195-3p. Results of bioinformatics analysis manifested that miR-5195-3p might have some complementary sites with JPX, as demonstrated by dual-luciferase reporter, RIP and RNA pull-down assay. Synchronously, our study also confirmed that miR-5195-3p expression level was inversely associated with JPX expression level in NSCLC cells. Then, we further probed whether the effect of JPX on NSCLC development was mediated by regulating miR-5195-3p. Functionally, JPX boosted proliferation, EMT and repressed apoptosis of NSCLC cells, and reintroduction of miR-5195-3p partly abolish the carcinogenic effect of JPX. In this paper, our study was the first to validate that JPX worked as an oncogenic factor in NSCLC by hindering the expression of miR-5195-3p.
VEGFA, as a member of the VEGF growth factor family, has been pointed out to be closely linked with cell growth and invasion in NSCLC.24 In the current study, we observed VEGFA was highly expressed in NSCLC tissues and cell lines. Interestingly, the negative relation between VEGFA and miR-5195-3p was observed. As widely believed, miRNAs could exert the role through the interaction of mRNA.31 In our study, results of bioinformatics analysis, dual-luciferase reporter and RIP assay verified the interaction between VEGFA and miR-5195-3p in NSCLC. In other words, miR-5195-3p could directly target VEGFA and regulate its expression in NSCLC cells. Besides, we viewed the positive correlation between VEGFA and JPX in NSCLC tissues. Notably, miR-5195-3p upregulation prominently suppressed the protein level of VEGFA and introduction of JPX partly reversed the inhibitory effect in NSCLC cells, implicating that JPX could serve as ceRNA of miR-5195-3p to affect the expression of VEGFA. Additionally, we executed rescue assays to prove that regulation of miR-5195-3p on NSCLC development was mediated by regulating VEGFA. As expected, miR-5195-3p impeded proliferation, EMT and accelerated apoptosis of NSCLC cells, and co-transfection of VEGFA partially overturned tumor-suppressive effect of miR-5195-3p.
Conclusions
Taken together, our study is the first to reveal the role of the JPX/miR-5195-3p/VEGFA axis in NSCLC development and progression, highlighting the potential of JPX as a new therapeutic target for NSCLC.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Johnson DH, Schiller JH, Bunn PA. Recent clinical advances in lung cancer management. J Clin Oncol. 2014;32(10):973–982. doi:10.1200/jco.2013.53.1228
3. Wood SL, Pernemalm M, Crosbie PA, Whetton AD. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev. 2015;41(4):361–375. doi:10.1016/j.ctrv.2015.02.008
4. Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64. doi:10.1210/er.2014-1034
5. Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015;3:5. doi:10.1186/s40591-015-0042-6
6. Wu R, Su Y, Wu H, Dai Y, Zhao M, Lu Q. Characters, functions and clinical perspectives of long non-coding RNAs. Mol Genet Genomics. 2016;291(3):1013–1033. doi:10.1007/s00438-016-1179-y
7. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi:10.1186/1476-4598-10-38
8. Dong Y, Huo X, Sun R, Liu Z, Huang M, Yang S. LncRNA Gm15290 promotes cell proliferation and invasion in non-small cell lung cancer through directly interacting with and suppressing the tumor suppressor miR-615-5p. Oncol Res. 2017. doi:10.3727/096504017x14930316817366
9. Miao F, Chen J, Shi M, Song Y, Chen Z. LncRNA HAND2-AS1 inhibits non-small cell lung cancer migration, invasion and maintains cell stemness through the interactions with TGF-beta1. Biosci Rep. 2019;39(1). doi:10.1042/bsr20181525
10. Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. doi:10.1016/j.cell.2010.09.049
11. Carmona S, Lin B, Chou T. LncRNA Jpx induces Xist expression in mice using both trans and cis mechanisms. PLoS Genetics. 2018;14(5):e1007378. doi:10.1371/journal.pgen.1007378
12. Jin M, Ren J, Luo M, et al. Long noncoding RNA JPX correlates with poor prognosis and tumor progression in non-small cell lung cancer by interacting with miR-145-5p and CCND2. Carcinogenesis. 2019. doi:10.1093/carcin/bgz125
13. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi:10.1016/j.addr.2015.05.001
14. Li S, Zeng X, Ma R, Wang L. MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp Ther Med. 2018;16(3):2038–2045. doi:10.3892/etm.2018.6370
15. Yu T, Li J, Yan M, et al. MicroRNA-193a-3p and −5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34(4):413–423. doi:10.1038/onc.2013.574
16. Song L, Dai Z, Zhang S, et al. MicroRNA-1179 suppresses cell growth and invasion by targeting sperm-associated antigen 5-mediated Akt signaling in human non-small cell lung cancer. Biochem Biophys Res Commun. 2018;504(1):164–170. doi:10.1016/j.bbrc.2018.08.149
17. Ebrahimi SO, Reiisi S. Downregulation of miR-4443 and miR-5195-3p in ovarian cancer tissue contributes to metastasis and tumorigenesis. Arch Gynecol Obstet. 2019;299(5):1453–1458. doi:10.1007/s00404-019-05107-x
18. Jiang Z, Zhang Y, Cao R, et al. miR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol Res. 2017;25(7):1081–1087. doi:10.3727/096504016x14831120463349
19. Yang Q. MicroRNA-5195-3p plays a suppressive role in cell proliferation, migration and invasion by targeting MYO6 in human non-small cell lung cancer. Biosci Biotechnol Biochem. 2019;83(2):212–220. doi:10.1080/09168451.2018.1540288
20. Petrovic N. Targeting angiogenesis in cancer treatments: where do we stand? J Pharm Pharm Sci. 2016;19(2):226–238. doi:10.18433/jpps.v19i2.27608
21. Ho QT, Kuo CJ. Vascular endothelial growth factor: biology and therapeutic applications. Int J Biochem Cell Biol. 2007;39(7–8):1349–1357. doi:10.1016/j.biocel.2007.04.010
22. Zhang W, Zou C, Pan L, et al. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37(3):1123–1133. doi:10.1159/000430237
23. Lu Y, Qin T, Li J, et al. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther. 2017;24(9):386–392. doi:10.1038/cgt.2017.30
24. Gu A, Lu J, Wang W, Shi C, Han B, Yao M. Role of miR-497 in VEGF-A-mediated cancer cell growth and invasion in non-small cell lung cancer. Int J Biochem Cell Biol. 2016;70:118–125. doi:10.1016/j.biocel.2015.10.013
25. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
26. Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8(2):e53823. doi:10.1371/journal.pone.0053823
27. Lantuejoul S, Constantin B, Drabkin H, Brambilla C, Roche J, Brambilla E. Expression of VEGF, semaphorin SEMA3F, and their common receptors neuropilins NP1 and NP2 in preinvasive bronchial lesions, lung tumours, and cell lines. J Pathol. 2003;200(3):336–347. doi:10.1002/path.1367
28. Li S, Mei Z, Hu HB. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. Journal of Cellular Physiology. 2018;233(9):6679–6688. doi:10.1002/jcp.26325
29. Li S, Yang J, Xia Y, Fan Q, Yang KP. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res. 2018;26(2):289–296. doi:10.3727/096504017x15009404458675
30. Yin D, Lu X, Su J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17(1):92. doi:10.1186/s12943-018-0836-7
31. Yang C, Sun C, Liang X, Xie S, Huang J, Li D. Integrative analysis of microRNA and mRNA expression profiles in non-small-cell lung cancer. Cancer Gene Ther. 2016;23(4):90–97. doi:10.1038/cgt.2016.5
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.