Back to Journals » OncoTargets and Therapy » Volume 12
Long Non-Coding RNA HULC Promotes the Development of Breast Cancer Through Regulating LYPD1 Expression by Sponging miR-6754-5p
Authors Wang N, Zhong C, Fu M, Li L, Wang F, Lv P, Zhu M, Xiong Y, Mi H, Gu Y
Received 5 August 2019
Accepted for publication 15 November 2019
Published 5 December 2019 Volume 2019:12 Pages 10671—10679
DOI https://doi.org/10.2147/OTT.S226040
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Nan Wang,1 Chaochao Zhong,2 Mingti Fu,3 Lin Li,1 Fang Wang,1 Pengwei Lv,1 Mingzhi Zhu,1 Youyi Xiong,1 Hailong Mi,1 Yuanting Gu1
1Department of Breast Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, People’s Republic of China; 2Department of Emergency, The First People’s Hospital of Zhengzhou, Zhengzhou 450052, Henan, People’s Republic of China; 3Department of General Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, People’s Republic of China
Correspondence: Yuanting Gu
Department of Breast Surgery, The First Affiliated Hospital of Zhengzhou University, Erqi District, Zhengzhou 450052, People’s Republic of China
Email [email protected]
Introduction: Long non-coding RNAs (lncRNAs) were found to regulate many biological processes including cancer development, immunology and other diseases. LncRNA HULC was found to be oncogenes in many cancer progression. However, the role of HULC in the regulation of breast cancer remains unclear.
Methods: The expression of HULC and miR-6754-5p was examined by RT-PCR. Through knockdown of HULC, we found that the proliferation abilities coupled with migration and invasion abilities were significantly decreased. And also, we verified that overexpression of miR-6754-5p significantly decreased the proliferation ability of breast cancer cells.
Results: In this study, we found that lncRNA HULC was overexpressed in breast cancer tissues and cell lines compared to normal healthy breast tissues and normal breast cell line. Moreover, the high expression of HULC was associated with metastasis and malignancy of breast cancers. Mechanically, we found that HULC can bind to miR-6754-5p directly through complementary base pairing. Furthermore, we found that HULC regulates the expression of LYPD1 through sponging miR-6754-5p. Moreover, overexpression of LYPD1 can rescue the migration and invasion abilities of breast cancer cells decreased by knockdown of HULC or overexpression of miR-6754-5p.
Conclusion: Our study showed the role of HULC in promoting breast cancer development and explained the detailed molecular mechanisms.
Keywords: lncRNA HULC, breast cancer development, miR-6754-5p, LYPD1
Introduction
Breast cancer is the most common invasive cancer in women. Breast cancer may also occur in men or children, or in pregnant women, but it is very rare.1 Adolescent women’s breasts consist of fat, connective tissue and thousands of lobules that provide milk for breastfeeding. In cancer, the body’s cells divide and grow uncontrollably. It is excessive cell growth that causes cancer. Breast cancer usually begins in the inner layer of the duct, or in the lobules that supply milk, from which it can spread to other parts of the body.2 In recent years, the incidence of breast cancer in young women is increasing. Breast cancer has very strong heterogeneity and complexity, so it is very important to study the molecular mechanism of breast cancer development.3
Long non-coding RNAs (lncRNAs) were found to be longer than 200nt and had little or noncoding potential.4 In recent years, many studies have proved that lncRNAs can exert very important roles in many biological processes especially cancer development.5–7 Highly upregulated in liver cancer (HULC) was firstly identified in liver cancer and was very highly expressed in liver cancer cells.8,9 Besides, many studies have found that HULC can also act as oncogenes to promote many other cancer progressions, including ovarian carcinoma, prostate cancer, chronic myeloid leukemia and so on.10–12 However, the function of HULC in the regulation of breast cancer progression remains unclear.
Ly6/PLAUR domain-containing protein 1 (LYPD1) is also known as Lynx2, a member of Lynx family of neurotransmitter receptor-binding protein.13 LYPD1 was reported to participate in the regulation of ovarian cancer and can act as a novel prognostic marker for ovarian cancer.14 And also, LYPD1 was proved to be essential for the endothelial network formation in bioengineered tissue.15 Moreover, LYPD1 was indicated to be associated with breast cancer metastasis. However, the role of LYPD1 in the regulation of breast cancer development remains unclear.
In our study, we found that HULC was also highly expressed in breast cancers compared to normal healthy breast tissues. And also, decreased expression of HULC was associated with lower proliferation, migration and invasion abilities. Mechanically, we found that HULC can bind to miR-6754-5p and inhibit the expression of miR-6754-5p through forming complementary base pairing. Moreover, we found that miR-6754-5p can bind to LYPD1. Through sponging miR-6754-5p, HULC promoted the expression of LYPD1. Consistently, overexpression of miR-6754-5p or inhibiting of LYPD1 also decreased the proliferation, migration and invasion of breast cancer cells.
Materials and Cells
Samples and Cell Lines
Human breast cancer samples and paired adjacent healthy breast tissues were obtained from 60 breast cancer patients under surgery at The First Affiliated Hospital of Zhengzhou University. All samples were kept in liquid nitrogen before use. This study was approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University. All written informed consents were received from patients.
Cell Lines
Breast cancer cell lines: MCF-7, ZR-75-1, BT-20, MDA-MB-231 and one normal breast cell line: MCF-10A were obtained from American Type Culture Collection (ATCC, USA). MCF-7 and MDA-MB-231 cells were cultured in DMEM medium supplemented with 100 mg/mL streptomycin, 100 U/mL penicillin and 10% FBS; BT-20 cells and ZR-75-1 cells were cultured in RPMI-1640 medium supplemented with 100 mg/mL streptomycin, 100 U/mL penicillin and 10% FBS. The culture medium of MCF-10A cells was M-171 medium and supplemented with mammary epithelial growth factors (Invitrogen/Life Technologies, USA).6 All cells were cultured at 37°C in 5% CO2.
CCK8 Assay
MCF-7 cells and BT-20 cells were seed into 96-well plate to examine the proliferation abilities using CCK8 detection kit (7 sea biotech, Shanghai, China) according to the manufacturer’s instruction.
Quantitative Reverse Transcription-PCR (qRT-PCR)
Total RNAs were extracted from tissues or cells using Trizol reagent. Then, RNAs were reverse-transcribed into cDNA using PrimeScript RT reagent Kit (Promega, Madison, WI, USA) according to the instruction of the kit. Finally, qRT-PCR assay was performed using SYBR Green PCR Master Mix reagents (Takara) in 7300 Real-Time PCR System (Applied Biosystems).
Transwell Assay
Cells were seeded into the Matrigel-coated chamber (BD Biosciences, Cowley, United Kingdom). The upper chambers were added with serum-free medium while the lower chambers were added with 10%FBS containing DMEM. After 24 hrs, cells in the lower chamber were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet.
Statistical Analysis
A Student’s t-test was used to analyze the differences between the two groups. A one-way ANOVA followed by a Tukey’s post hoc test was used for multiple comparisons. The effect of lncRNA HULC expression on overall survival was analyzed by Kaplan-Meier analysis and a log-rank test. GraphPad Prism 6 software was used to analyze all results. The results were expressed as mean ± SD. P < 0.05 was considered to be significant.
Results
Elevated Expression of lncRNA HULC in Breast Cancer Tissues and Cell Lines Is Associated with Poor Prognosis
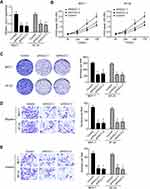
LncRNAs were proved to promote many cancers development as oncogenes. However, the role of HULC in breast cancer development remains unclear. So, we wanted to explore the function of HULC in breast cancer progression. Firstly, we found that the expression of HULC was remarkably higher in breast cancer cells, MCF-7, BT-20, ZR-75-1 and MDA-MB-231 relative to normal breast cell, MCF-10A (Figure 1A). Besides, we also collected 60 pairs of human breast cancer tissues and adjacent normal breast tissues. Through qRT-PCR analysis, we found that lncRNA HULC was very highly expressed in breast cancer tissues compared to adjacent healthy tissues (Figure 1B). Moreover, we also examined the expression of HULC in breast cancer tissues with metastasis and advanced stages, we observed that higher expression was associated with metastasis and advanced stages (Figure 1C–D). To further examine the overall survival rate of HULC, we performed Kaplan-Meier curve analysis. We divided the 60 samples into two groups based on the median expression of HULC (Table 1). Consistently, we found that lower HULC expression possessed better overall survival (Figure 1E). Taken together, we observed that HULC expression was upregulated in breast cancer tissues and cell lines and associated with poor overall survival rate.
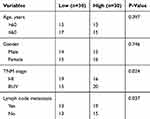 |
Table 1 Correlation Between HULC Expression and Clinicopathological Features in 60 Patients with Breast Cancer |
Knockdown of HULC Expression Significantly Impairs Development of Breast Cancer
As we showed above, we found that HULC was overexpressed in breast cancer tissues and cell lines. To explore the function of HULC in the regulation of breast cancer development, we constructed shRNAs of HULC (Figure 2A). Firstly, we examined the proliferation abilities of breast cancer cells using CCK8 assay after knockdown of HULC. We found that decreased HULC expression leads to decreased proliferation of MCF-7 cells and BT-20 cells (Figure 2B). And, we also performed colony formation assay. Consistently, we observed that knockdown of HULC decreased the numbers of colonies when compared to the negative control (Figure 2C). As the expression of HULC was higher in metastasis and advanced breast cancer tissues, so we wanted to explore if HULC regulated the migration and invasion of MCF-7 cells and BT-20 cells. To examine that, we performed migration and invasion assay and observed significantly less numbers of migration and invasion cells after knockdown of HULC (Figure 2D–E). Collectively, we found that HULC promoted the development of MCF-7 cells and BT-20 cells.
miR-6754-5p Is Sponged by HULC and Regulates the Expression of HULC
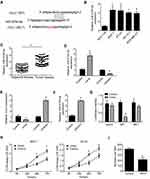
To find the potential mechanism that HULC regulated the development of breast cancer, we performed bioinformatics analysis using HULC sequence. We found that HULC can form complementary base pairing with miR-6754-5p directly (Figure 3A). Moreover, we found that the expression of miR-6754-5p was elevated in breast cancer cells compared to normal breast cells (Figure 3B). And also, miR-6754-5p was highly expressed in breast cancer tissues compared to adjacent tissues (Figure 3C). To explore the regulation between HULC and miR-6754-5p, we constructed miR-6754-5p mimic and inhibitor (Figure 3D). Through transfection and qRT-PCR analysis, we observed that overexpression of miR-6754-5p significantly decreased HULC expression while inhibition of miR-6754-5p remarkably increased the expression of HULC (Figure 3E). Moreover, we also found that knockdown of HULC increased miR-6754-5p expression (Figure 3F). To validate the interaction between HULC and miR-6754-5p, we performed luciferase reporter assay. We observed that the luciferase intensity of WT-HULC was decreased after ectopic expression of miR-6754-5p while the luciferase intensity of MUT-HULC did not change (Figure 3G). Finally, we also wanted to explore if miR-6754-5p regulated the proliferation of breast cancer cells. We performed CCK8 assay and found that miR-6754-5p remarkably decreased proliferation abilities of MCF-7 cells and BT-20 cells examined by CCK8 assay and colony formation assay (Figure 3H–I). In sum, we found that HULC interacts with miR-6754-5p and miR-6754-5p can inhibit the progression of breast cancer.
LYPD1 Is a Potential Target for HULC and miR-6754-5p in the Regulation of Breast Cancer Progression
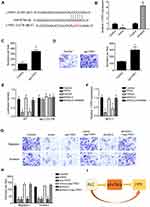
To find the potential target for HULC and miR-6754-5p, we performed TargetScan7 analysis. We found that miR-6754-5p can bind to LYPD1 directly (Figure 4A). Furthermore, through qRT-PCR assay, we observed that overexpression of miR-6754-5p decreased the expression of LYPD1 while inhibition of miR-6754-5p increased LYPD1 expression significantly (Figure 4B). Then, we found that overexpression of LYPD1 could significantly increase the proliferation abilities and migration abilities (Figure 4C-D). Moreover, to validate the interaction between miR-6754-5p and LYPD1, we performed luciferase assay and proved that ectopic expression of miR-6754-5p or knockdown of HULC significantly decreased the luciferase intensity of WT-HULC. And also, inhibition of miR-6754-5p coupled with knockdown of HULC can rescue the luciferase intensity of WT-HULC decreased by knockdown of HULC (Figure 4E). Moreover, the expression level of LYD1 was consistent with the luciferase intensity of WT-HULC (Figure 4F). To validate the relationship of HULC, miR-6754-5p and LYPD1 in regulating breast cancer development, we performed migration and invasion assay. We observed that ectopic expression of miR-6754-5p, knockdown of HULC or knockdown of LYPD1 significantly decreased the migration and invasion of breast cancer cells while overexpression of miR-6754-5p after overexpression of LYPD1 or overexpression of LYPD1 after knockdown of HULC can rescue the decreased migration and invasion of breast cancer cells (Figure 4G–H). Collectively, we showed that lncRNA HULC promoted breast cancer development through miR-6754-5p/LYPD1 axis (Figure 4I).
Discussion
Breast cancer is a common malignant tumor in women, which seriously threatens women’s physical and mental health.16 The etiology of breast cancer is not completely clear.17 Moreover, studies have found that there are certain regularities in the occurrence of breast cancer.18 So, it is important to discover the potential mechanisms that regulate the progression of breast cancer.
LncRNAs were proved to play vital functions in many cancer progression including breast cancer.19,20 For example, Shi SJ and colleagues found that lncRNA-ATB can promote the invasion and metastasis cascade of breast cancer.21 And also, Li Z and colleagues proved that lncRNA ANCR impairs the migration and invasion of breast cancer cells through the degradation of EZH2.22 Though there are many researches focusing on the regulation of breast cancer by lncRNAs, there are still some problems need to be solved.
lncRNA HULC is firstly identified in liver cancer. It was proved that the expression of lncRNA HULC is significantly elevated in liver cancer compared to normal liver tissues.23–25 And also, lncRNA HULC was proved to promote the progression of liver cancer.26,27 Besides, there are other evidences that proved lncRNA HULC participates in the regulation of other cancers.28–30 Chen S and colleagues observed that lncRNA HULC participates in the regulation of epithelial ovarian carcinoma by targeting ATG7 and ITGB1.31 And also, Kong D and colleagues discovered that lncRNA HULC promotes apoptosis of osteosarcoma cells and inhibits the proliferation, migration and invasion of osteosarcoma cells by sponging miR-122.32 However, the role of lncRNA HULC in regulating breast cancer remains unknown.
In this study, we found that lncRNA HULC is highly expressed in breast cancer tissues and cell lines. Moreover, we discovered that HULC promotes breast cancer cells’ proliferation, migration and invasion by sponging miR-6754-5p. miR-6754-5p was reported to inhibit the progression of non-small lung cancer through regulated by lncRNA LINC00978.33 In our study, we firstly reported the role of miR-6754-5p in the regulation of breast cancer. Furthermore, the functions of miR-6754-5p in regulating other cancers still need to be explored.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ennour-Idrissi K, Ayotte P, Diorio C. Persistent organic pollutants and breast cancer: a systematic review and critical appraisal of the literature. Cancers (Basel). 2019;11. doi:10.3390/cancers11081063
2. De Cicco P, Catani MV, Gasperi V, Sibilano M, Quaglietta M, Savini I. Nutrition and breast cancer: a literature review on prevention, treatment and recurrence. Nutrients. 2019;11. doi:10.3390/nu11071514
3. Gardezi SJS, Elazab A, Lei B, Wang T. Breast cancer detection and diagnosis using mammographic data: systematic review. J Med Internet Res. 2019;21:e14464. doi:10.2196/14464
4. Guo CJ, Zhang W, Gershwin ME. Long noncoding RNA lncKdm2b: a critical player in the maintenance of group 3 innate lymphoid cells. Cell Mol Immunol. 2018;15:5–7. doi:10.1038/cmi.2017.55
5. Fu J, Dong G, Shi H, et al. LncRNA MIR503HG inhibits cell migration and invasion via miR-103/OLFM4 axis in triple negative breast cancer. J Cell Mol Med. 2019;23:4738–4745. doi:10.1111/jcmm.14344
6. Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. 2018;495:1594–1600. doi:10.1016/j.bbrc.2017.12.013
7. Kong X, Duan Y, Sang Y, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105–9117. doi:10.1002/jcp.v234.6
8. Yu X, Zheng H, Chan MT, Wu WK. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med. 2017;21:410–417. doi:10.1111/jcmm.12956
9. Wang BG, Lv Z, Ding HX, et al. The association of lncRNA-HULC polymorphisms with hepatocellular cancer risk and prognosis. Gene. 2018;670:148–154. doi:10.1016/j.gene.2018.05.096
10. Feng H, Wei B, Zhang Y. Long non-coding RNA HULC promotes proliferation, migration and invasion of pancreatic cancer cells by down-regulating microRNA-15a. Int J Biol Macromol. 2019;126:891–898. doi:10.1016/j.ijbiomac.2018.12.238
11. Wang YF, Zhang S, Li XQ, Wang Y. Expression of lncRNA HULC in cervical cancer and its correlation with tumor progression and patient survival. Eur Rev Med Pharmacol Sci. 2016;20:3987–3991.
12. Wang J, Ma W, Liu Y. Long non-coding RNA HULC promotes bladder cancer cells proliferation but inhibits apoptosis via regulation of ZIC2 and PI3K/AKT signaling pathway. Cancer Biomark. 2017;20:425–434. doi:10.3233/CBM-170188
13. Egerod KL, Holst B, Petersen PS, et al. GPR39 splice variants versus antisense gene LYPD1: expression and regulation in gastrointestinal tract, endocrine pancreas, liver, and white adipose tissue. Mol Endocrinol. 2007;21:1685–1698. doi:10.1210/me.2007-0055
14. Sandow JJ, Rainczuk A, Infusini G, et al. Discovery and validation of novel protein biomarkers in ovarian cancer patient urine. Proteomics Clin Appl. 2018;12:e1700135. doi:10.1002/prca.v12.3
15. Masuda S, Matsuura K, Shimizu T. Inhibition of LYPD1 is critical for endothelial network formation in bioengineered tissue with human cardiac fibroblasts. Biomaterials. 2018;166:109–121. doi:10.1016/j.biomaterials.2018.03.002
16. Guerra RL, Castaneda L, de Albuquerque RCR, et al. Patient preferences for breast cancer treatment interventions: a systematic review of discrete choice experiments. Patient. 2019. doi:10.1007/s40271-019-00375-w
17. Savva C, Adhikaree J, Madhusudan S, Chokkalingam K. Oncogenic osteomalacia and metastatic breast cancer: a case report and review of the literature. J Diabetes Metab Disord. 2019;18:267–272. doi:10.1007/s40200-019-00398-y
18. Farahmand M, Monavari SH, Shoja Z, Ghaffari H, Tavakoli M, Tavakoli A. Epstein-barr virus and risk of breast cancer: a systematic review and meta-analysis. Future Oncol. 2019. doi:10.2217/fon-2019-0232
19. Guan Y, Bhandari A, Xia E, Yang F, Xiang J, Wang O. lncRNA FOXD3-AS1 is associated with clinical progression and regulates cell migration and invasion in breast cancer. Cell Biochem Funct. 2019;37:239–244. doi:10.1002/cbf.v37.4
20. Liu H, Deng ZH, Han F, Xia YY, Liu QH, Sato M. Time-frequency analysis of air-coupled GPR data for identification of delamination between pavement layers. Constr Build Mater. 2017;154:1207–1215. doi:10.1016/j.conbuildmat.2017.06.132
21. Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi:10.18632/oncotarget.3457
22. Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59–71. doi:10.1038/cdd.2016.95
23. Shen X, Guo H, Xu J, Wang J. Inhibition of lncRNA HULC improves hepatic fibrosis and hepatocyte apoptosis by inhibiting the MAPK signaling pathway in rats with nonalcoholic fatty liver disease. J Cell Physiol. 2019;234:18169–18179. doi:10.1002/jcp.28450
24. Wang Y, Chen F, Zhao M, et al. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J Biol Chem. 2017;292:15395–15407. doi:10.1074/jbc.M117.783738
25. Du Y, Kong G, You X, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi:10.1074/jbc.M112.342113
26. Hammerle M, Gutschner T, Uckelmann H, et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology. 2013;58:1703–1712. doi:10.1002/hep.26537
27. Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi:10.1093/nar/gkq285
28. Zhong Y, Chen Z, Guo S, et al. TUG1, SPRY4-IT1, and HULC as valuable prognostic biomarkers of survival in cancer: a PRISMA-compliant meta-analysis. Medicine (Baltimore). 2017;96:e8583. doi:10.1097/MD.0000000000008583
29. Lu D, Wei HX, Zhang J, et al. Part 1: notch-sparing gamma-secretase inhibitors: the identification of novel naphthyl and benzofuranyl amide analogs. Bioorg Med Chem Lett. 2016;26:2129–2132. doi:10.1016/j.bmcl.2016.03.040
30. Kang M, Sang Y, Gu H, et al. Long noncoding RNAs POLR2E rs3787016 C/T and HULC rs7763881 A/C polymorphisms are associated with decreased risk of esophageal cancer. Tumour Biol. 2015;36:6401–6408. doi:10.1007/s13277-015-3328-z
31. Chen S, Wu DD, Sang XB, et al. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017;8:e3118.
32. Kong D, Wang Y. Knockdown of lncRNA HULC inhibits proliferation, migration, invasion, and promotes apoptosis by sponging miR-122 in osteosarcoma. J Cell Biochem. 2018;119:1050–1061. doi:10.1002/jcb.26273
33. Li X, Ren Y, Zuo T. Long noncoding RNA LINC00978 promotes cell proliferation and invasion in nonsmall cell lung cancer by inhibiting miR67545p. Mol Med Rep. 2018;18:4725–4732. doi:10.3892/mmr.2018.9463
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.