Back to Journals » OncoTargets and Therapy » Volume 12
Long Non-Coding RNA CASC9 And HIF-1α Form A Positive Feedback Loop To Facilitate Cell Proliferation And Metastasis In Lung Cancer
Authors Jin Y, Xie H, Duan L, Zhao D, Ding J, Jiang G
Received 5 August 2019
Accepted for publication 21 October 2019
Published 31 October 2019 Volume 2019:12 Pages 9017—9027
DOI https://doi.org/10.2147/OTT.S226078
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Yuxing Jin, Huikang Xie, Liang Duan, Deping Zhao, Jiaan Ding, Gening Jiang
Department of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai 200433, People’s Republic of China
Correspondence: Gening Jiang
Department of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai 200433, People’s Republic of China
Email [email protected]
Background: The long noncoding RNA cancer susceptibility 9 (CASC9) has been recognized as an important modulator of cell growth and metastasis in many cancers. However, its detailed roles in lung cancer remain unclear. In this study, we aimed to investigate its functions and molecular mechanism in lung cancer progression.
Methods: Expression of CASC9 was determined in lung cancer tissues and cell lines by real-time PCR. CCK-8, colony formation, wound healing and transwell assays were done to evaluate the cell proliferation, migration and invasion capacities in vitro. Real-time PCR, Western blot and RNA immunoprecipitation (RIP) assays were performed to dissect the mechanisms.
Results: CASC9 was overexpressed in lung cancer specimens and cell lines. Knockdown of CASC9 inhibited cell proliferation, migration, invasion and EMT in lung cancer cells. While overexpression of CASC9 in normal lung epithelial cells did the opposite. CASC9 interacted with HIF-1α and enhanced its protein stability. They formed a positive feedback loop by reciprocally inducing each other expression and regulated cell proliferation and metastasis.
Conclusion: Our findings demonstrated a novel regulatory signaling pathway, namely the CASC9/HIF-1α axis, which was involved in lung cancer progression. These findings can provide valuable insights on the potential therapy application for lung cancer.
Keywords: CASC9, HIF-1α, cell proliferation, metastasis, lung cancer
Introduction
Lung cancer is one of the frequently diagnosed and the leading cause of cancer-related death in the world. An estimated 1.8 million new lung cancer cases occurred in 2012, accounting for about 13% of total cancer diagnoses.1 Considering its high incidence and mortality, intensive investigations are required to learn more about the detailed molecular mechanisms governing the initiation and progression of lung cancer. However, the current knowledge about this disease remain limited, and the prognosis stay largely unsatisfactory.
Long non-coding RNAs (lncRNAs) are important regulatory factors in cancer development and progression. They are a group of RNAs with over 200 nucleotides in length and with no protein-coding potential.2 In decades, non-coding RNAs (ncRNAs) are considered as the transcriptional byproducts without biological roles on pathophysiological processes. With the help of next‐generation sequencing (NGS) and bioinformatics tools, the crucial roles of lncRNAs have been wildly recognized recently.3,4
Cancer susceptibility 9 (CASC9) was first defined upregulated lncRNA in esophageal squamous cell carcinoma (ESCC) by RNA sequencing, and knockdown of CASC9 significantly suppressed cell migration and invasion in ESCC cells.5 It has also been reported to promote tumor progression in several other cancers such as ovarian cancer, esophageal cancer, glioma, lung cancer and liver cancer.5–10 Although it has been mentioned that CASC9 could promote cell proliferation and metastasis in lung cancer,9 the detailed mechanism remains unclear.
In our study, we intended to uncover the role and mechanism of CASC9 in lung cancer. We found that CASC9 formed a positive feedback loop with HIF-1α and promoted cell proliferation and metastasis in lung cancer. In summary, our findings revealed a novel regulatory signaling pathway, namely the CASC9/HIF-1α axis, involved in lung cancer progression. These findings can provide valuable insights on the potential therapy application for lung cancer.
Materials And Methods
Collection Of Lung Cancer Specimens
Lung cancer specimens were collected from 40 patients who underwent surgery resection at the Department of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine between 2012 and 2016. Clinical information was collected from medical records. The 40 patients ranged in age from 48 to 74. Histological diagnosis was accomplished by two pathologists in a double-blind manner. After specimens obtaining, they were snap-frozen by liquid nitrogen saved at −80°C until used. This study was approved by the Ethics Committee of Shanghai Pulmonary Hospital, Tongji University School of Medicine and each patient signed the written informed consent before enrollment in the study.
Cell Culture And Treatment
Human normal lung epithelial cell line HFL-1, and lung cancer cell lines 95D, A549, PC-9, SPC-A1 and HAC-84 were purchased from Chinese Academy of Science. HFL-1 cells were cultured in F-12K medium containing 10% fetal bovine serum (FBS), other cell lines were maintained in RMPI-1640 medium supplemented with 10% FBS. All cells were incubated at 37°C with 5% CO2.
Molidustat (10 μM; Selleck, China) was used to treat cells for 4h to mimic HIF-1α activation. BAY87-2243 (10 μM; Selleck, China) was used to treat cells for 48h to induce HIF-1 inhibition. Proteasome inhibitor MG132 (25 μM; MedChem Express, China) and protein translation inhibitor puromycin (2 μM; Sigma, USA) were also used to treat cells for indicated time. Then cells were harvested for Western blot or real-time PCR.
RNA Knockdown By Small Interfering RNA
Small interfering RNA (siRNA) targeting CASC9 or the scrambled oligonucleotides used as negative control (NC) were synthesized by Gene Pharma (China). The sequences of siRNAs were as follows: NC sense: 5ʹ-UUC UCC GAA CGU GUC ACG UTT-3ʹ, NC antisense: 5ʹ-ACG UGA CAC GUU CGG AGA ATT-3ʹ, si-CASC9 sense: 5ʹ-GGA CUC AUA UUA CCA GUC UTT-3ʹ, si-CASC9 antisense: 5ʹ-AGA CUG GUA AUA UGA GUC CTT-3ʹ. For siRNA transfection, cells were pre-seeded in six-well plates at a density of 3×105/well overnight, and then transfected with si-CASC9 or NC at a final concentration of 100 nM using Lipofectamine 2000 (Invitrogen, USA). 72 h later, RNA was extracted, and the interfering efficiency was determined by real-time PCR.
Overexpression Of CASC9 By Plasmid Transfection
CASC9 expresssing plasmid was synthesized by Gene Pharma (China) in pcDNA3.1 vector. To overexpress CASC9 in HFL-1 cells, cells were pre-seeded in six-well plates at a density of 5×105/well and settled down overnight. Transfection was accomplished by using Lipofectamine 3000 (Invitrogen, USA) and 2 μg plasmid (empty pcDNA3.1 vector as the control and CASC9 expressing plasmid) for each well. 24 h later, RNA was extracted, and the overexpression efficiency was determined by real-time PCR.
CCK-8, Colony Formation Assay For Cell Proliferation
For CCK-8 assay, cells pre-transfected with CASC9 siRNA for 48h were plated in 96-well plates at 3000 cells per well and incubated overnight. CCK-8 reagent was added into the wells. After incubating at 37°C for another 2 h, absorbance was measured at 450 nm using a microplate reader. For colony formation assay, cells pre-transfected with CASC9 siRNA for 48h were plated in 6-well plates at 2000 cells per well and incubated for another 2 weeks. Then cells were fixed by methanol, stained by crystal violet solution, and photographed under an inverted microscope. All experiments were undertaken three times and data were expressed as mean ± SD.
Wound Healing Assay
For the wound healing assay, a 200 μL pipette tip was used to obtain the cell free lane. A ruler was used as a guide to obtain a straight line. To remove the detached cells, the culture medium was discarded, and the cells were washed twice with phosphate-buffered saline (PBS). Cells were continued to be cultured and allowed to migrate for 48 h. Distances between the edges of the scratch were measured to quantitatively evaluate cell migration.
Transwell Migration And Invasion Assays
Transwell migration and invasion assays were performed by using Millicell inserts (Corning) pre-coated without and with Matrigel, respectively. 2.5×104 cells were seeded per upper chambers in serum-free 1640 (for 95D and A549 cells) or F-12K (for HFL-1 cells) whereas the lower chambers were loaded with the same 1640 or F-12K medium containing 5% FBS. After 24 h, the non-migrated or non-invaded cells on the upper chambers were removed by a cotton swab, and cells migrated or invaded to the underside of the membrane were stained and counted.
Western Blot
Equal amounts of protein lysates were separated by SDS-PAGE and transferred onto PVDF membranes. Filters were probed with the following primary antibodies: anti-HIF-1α, anti-E-cadherin, anti-N-cadherin, anti-Vimentin, anti-β-catenin and anti-β-actin (Cell Signaling Technology, USA). Blots were then incubated with horseradish peroxidase-conjugated secondary antibody (Pierce, USA) and visualized by chemi-luminescence.
Real-Time PCR
Total RNAs were extracted from tissues or cells using the TRIZOl reagent (Invitrogen, USA) according to the manufacturer’s protocol. Real-time PCR was done with the SYBR Green PCR kit (Takara Bio, China) to determine the target mRNA expression in this study. The sequences of real-time PCR primers used were as follows: CASC9 sense: 5ʹ-AGA TGA AGC CGG TAC CTC AGA T-3ʹ; CASC9 antisense: 5ʹ-TCA CTT TAA AGA GGG AGA GGA G-3ʹ; HIF-1α sense: 5ʹ-AGT CGG ACA GCC TCA CCA A-3ʹ; HIF-1α antisense: 5ʹ-TGC TCC ATT CCA TTC TGT TCA-3ʹ; VEGF sense: 5ʹ-GGA AGA GTA GCT CGC CGA GG-3ʹ; VEGF antisense: 5ʹ-GGA GGT AGA GCA GCA AGG CA-3ʹ; GLUT1 sense: 5ʹ-CAT CCT CAT CGC CCA GGT GT-3ʹ; GLUT1 antisense: 5ʹ-GCG GTT GAT GAG CAG GAA GC-3ʹ; E-cadherin sense: 5ʹ-AGT CAC TGA CAC CAA CGA TAA T-3ʹ; E-cadherin antisense: 5ʹ-ATC GTT GTT CAC TGG ATT TGT G-3ʹ; N-cadherin sense: 5ʹ-CGA TAA GGA TCA ACC CCA TAC A-3ʹ; N-cadherin antisense: 5ʹ-TTC AAA GTC GAT TGG TTT GAC C-3ʹ; Vimentin sense: 5ʹ-TTG CCG TTG AAG CTG CTA ACT ACC-3ʹ; Vimentin antisense: 5ʹ-AAT CCT GCT CTC CTC GCC TTC C-3ʹ; GAPDH sense: 5ʹ-CCA CCC ATG GCA AAT TCC ATG GCA-3ʹ; GAPDH antisense: 5ʹ-TCT AGA CGG CAG GTC AGG TCC ACC-3ʹ. Relative mRNA expression was normalized to GAPDH and calculated according to the 2−ΔΔCt method.
RNA Immunoprecipitation (RIP) Assay
CASC9 RNA was firstly conjugated with biotin and incubated with purified His-HIF-1α protein for 4 h at room temperature. Streptavidin agarose beads were then added into the above complex and rotated overnight at 4°C. The beads were collected and resuspended in SDS-PAGE loading buffer followed by four times of washing with cold PBS. After denaturation of the precipitate, the interaction of CASC9 RNA with HIF-1α was visualized by Western blot.
Statistical Analysis
Data were collected from at least three independent experiments, with three replicates performed per experiment. All data are expressed as the mean ± SD. Statistically significant effects (*P<0.05, **P<0.01, ***P<0.001) were evaluated by a two-tailed Student’s t-test.
Results
CASC9 Is Overexpressed In Lung Cancer Tissues And Cell Lines
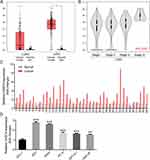
To understand the role of CASC9 in lung cancer, we firstly predicted its expression status in web-based lung cancer tissues from the GEPIA dataset (Gene Expression Profiling Interactive Analysis; http://gepia.cancer-pku.cn). The results showed that CASC9 was significantly overexpressed in LUSC (lung squamous cell carcinoma) tissues, while slightly increased in LUAD (lung adenocarcinoma) tissues compared with their respective normal controls (Figure 1A). Specifically, CASC9 expression was positively correlated with the advanced tumor stage of LUSC tissues. Patients in stage IV LUSC probably contained the highest level of CASC9 (Figure 1B). The overexpression of CASC9 was further proved in 40 pairs of lung cancer tissues matched with their adjacent normal tissues by real-time PCR (Figure 1C). Also, compared with that in human normal lung epithelial HFL-1 cells, CASC9 expression was obviously increased in lung cancer cell lines, in which 95D and A549 exhibited higher CASC9 levels (Figure 1D). These results imply that CASC9 may act as an important factor in lung cancer progression.
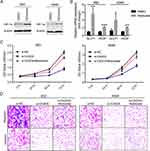
Knockdown Of CASC9 Inhibits Lung Cancer Cell Proliferation
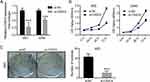
From the above expression detection results, we observed that CASC9 was highly expressed in 95D and A549 cell lines, then we performed CASC9 knockdown assay in the two cell lines by transfection of CASC9 specific siRNA (si-CASC9). As shown in Figure 2A, transfection of si-CASC9 significantly decreased the expression of CASC9 in 95D and A549 cell. Then CCK-8 assay was used to measure cell proliferation alteration by CASC9 knockdown. As shown in Figure 2B, knockdown of CASC9 significantly inhibited cell proliferation in the two lung cancer cell lines. Colony formation assay also revealed that CASC9 knockdown resulted in growth inhibition in 95D cells (Figure 2C), indicating that CASC9 itself promotes lung cancer cell proliferation.
Knockdown Of CASC9 Inhibits Lung Cancer Cell Migration, Invasion And EMT
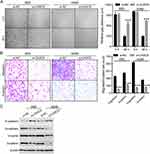
As CASC9 was shown to promote cancer progression by affecting epithelial-mesenchymal transition (EMT) procedure,5,7,9,11 we wondered whether CASC9 could also promote migration, invasion and EMT in lung cancer cells. To test this, wound healing assay was performed to evaluate cell migration ability upon CASC9 knockdown. The results showed that CASC9 knockdown significantly inhibited cell migration in 95D and A549 cells (Figure 3A). Transwell migration and invasion assays showed similar results (Figure 3B), illustrating that CASC9 promotes cell migration and invasion in lung cancer.
EMT is a process by which epithelial cells lose their cell polarity and cell-cell adhesion and gain migratory and invasive properties to become mesenchymal cells.12 EMT also occurs in the initiation of metastasis in cancer progression. As we demonstrated that knockdown of CASC9 significantly inhibited cell migration and invasion in lung cancer cells, we suspected that CASC9 could promote EMT procedure in lung cancer. To verify this hypothesis, we measured the expression of E-cadherin, N-cadherin, Vimentin and β-catenin, which can be used as markers for EMT.13 Western blot showed that knockdown of CASC9 significantly increased the protein level of E-cadherin and decreased the protein levels of N-cadherin, Vimentin and β-catenin (Figure 3C), indicating that knockdown of CASC9 inhibit EMT.
Overexpression Of CASC9 In Normal HFL-1 Cells Promotes Cell Proliferation, Migration And Invasion
To fully characterize the role of CASC9, we also performed gain-of-function studies in the normal HFL-1 cells, since it expressed the lowest endogenous level of CASC9 (Figure 1D). Overexpression was finished by plasmid transfection and confirmed by real-time PCR (Supplementary Figure 1A). Then CCK-8 and clony formation assays supported that CASC9 promoted HFL-1 cell proliferation (Supplementary Figure 1B and C). Additionally, it also facilitated HFL-1 cell migration and invasion obviously (Supplementary Figure 1D and E). These results further emphasized the oncogenic roles of CASC9 in lung cancer.
CASC9 Binds To HIF-1α And Enhances Its Stability
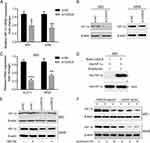
It is well recognized that hypoxia-inducible factor 1 alpha (HIF-1α) is involved in cancer metastasis through many signal pathways.14–16 CASC9 was reported to interact with HIF-1α and stimulated its downstream targets.17 Herein, we found that knockdown of CASC9 not only reduced mRNA level of HIF-1α but also decreased the protein level of HIF-1α (Figure 4A and B), suggesting that CASC9 may regulate lung cancer cell metastasis though affecting the HIF-1α level. In addition, CASC9 knockdown also led to decreased expression of GLUT1 and VEGF, the two well-known direct targets of HIF-1α (Figure 4C). As CASC9 has been reported to interact and stabilize HIF-1α protein in nasopharyngeal carcinogenesis, we wonder whether it could function similarly in lung cancer. As shown in Figure 4D, RIP assay revealed the interaction of CASC9 RNA with HIF-1α protein. MG132 treatment could nearly recover the protein level of HIF-1α protein to the extent of the control group (Figure 4E). However, after treatment of puromycin, which can block protein synthesis, HIF-1α protein was more stable in CASC9-overexpressed cells (Figure 4F), demonstrating that CASC9 was mainly involved in the proteostasis control of HIF-1α.
Restoration Of HIF-1α Recovers Lung Cancer Proliferation And Metastasis Defects Induced By CASC9 Knockdown
From the above results, we established that CASC9 was a crucial oncogenic lncRNA in lung cancer cells by promoting cell proliferation, migration, invasion and EMT. The control of HIF-1α proteostasis might be implicated in the tumor facilitating effects of CASC9. Then we questioned whether HIF-1α restoration could rescue the observed proliferation and metastasis defects caused by CASC9 knockdown. To this end, Molidustat, the HIF-stabilizer, was used to treat CASC9-silenced 95D and A549 cells to mimic hypoxia. Real-time PCR validated the successful induction of HIF-1α protein and GLUT1 and VEGF gene expression upon Molidustat treatment (Figure 5A and B). CCK-8, wound healing and transwell invasion assays in 95D and A549 cells all confirmed that Molidustat combined with si-CASC9 treatment significantly recovered lung cancer cell proliferation, migration and invasion to the extent of the control group, respectively (Figure 5C and D). These results suggested that CASC9 acted through HIF-1α to function as an oncogene in lung cancer progression.
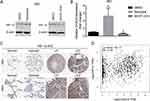
HIF-1α Positively Regulates CASC9 Expression
To test whether HIF-1α could affect CASC9 expression or not, we used Molidustat to stimulate HIF-1α expression and used BAY87-2243 to inhibit HIF-1α expression. As shown in Figure 6A, expression of HIF-1α protein dramatically increased after treatment with 10 uM Molidustat for 4h. Interestingly, the expression of CASC9 was also significantly increased (Figure 6B). Consistently, after inhibition of HIF-1α expression by treatment of 10 uM BAY87-2243 for 24h (Figure 6A), the expression of CASC9 was significantly decreased (Figure 6B). These results showed that HIF-1α can also positively regulate CASC9 expression, suggesting a positive feedback loop existed between CASC9 and HIF-1α. Finally, immunohistochemical analyses from the human protein atlas (https://www.proteinatlas.org) verified that HIF-1α was markedly overexpressed in lung cancer tissues, accumulating in both cytoplasm and nucleus (Figure 6C). And there was a significant positive correlation between HIF1A and CASC9 in lung cancer tissues (Figure 6D). Taken together, our results indicate a positive feedback loop between CASC9 and HIF-1α by reciprocally inducing each other expression, therefore favoring lung cancer cell proliferation and metastasis.
Discussion
Lung cancer was the most frequently diagnosed cancer and the leading cause of cancer death among the world population.1 Understanding the molecular mechanisms and pathways of lung cancer is conducive to enhance the efficacy of lung cancer treatment and improve the prognosis of lung cancer patients.
CASC9 was firstly identified to be upregulated in esophageal squamous cell carcinoma (ESCC).5 Then increasing lines of evidence show that CASC9 could promote cancer cell proliferation and metastasis in several other malignancies. In our study, we detected the expression of CASC9 in lung cancer tissues and cell lines firstly. Then loss-of-function assays were performed to knockdown CASC9 and measure the cell proliferation, migration, invasion and EMT activities in 95D and A549 lung cancer cells. Combining in silico analyses and experimental studies, we confirmed that CASC9 was indeed upregulated in lung cancer, which kept consistent with the previous report in lung adenocarcinoma.9 Functionally, knockdown of CASC9 in two lung cancer cell lines, 95D and A549 cell lines, who contained relatively higher level of endogenous CASC9, significantly inhibited cell proliferation, migration, invasion and EMT, suggesting that CASC9 itself could promote cell proliferation and metastasis in lung cancer cells. Consistently, when overexpressing CASC9 in the normal lung epithelial HFL-1 cells, it exhibited strong oncogenic activities to promote cell proliferation, migration and invasion. Since EMT is a process by which epithelial cells lose their cell polarity and cell-cell adhesion and gain migratory and invasive properties to become mesenchymal cells,12 it occurs in the initiation of metastasis in cancer progression. Therefore, we consider that the pro-migration and pro-invasion roles of CASC9 in lung cancer might result from its positive impact on EMT induction. Consistent with the results in the previous literature,9 our results showed that knockdown of CASC9 significantly increased the protein level of E-cadherin and decreased the protein levels of N-cadherin, Vimentin and β-catenin, indicating that CASC9 truly promoted EMT procedure.
According to the known mechanisms, CASC9 might promote tumor progression by affecting many signaling pathways. For instance, CASC9 can promote LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer.6 CASC9 can interact with CREB-binding protein to upregulate LAMC2 expression and promotes esophageal squamous cell carcinoma metastasis.7 CASC9 promotes nasopharyngeal carcinogenesis via stabilizing HIF-1α.17 CASC9 forms a complex with HNRNPL to regulate AKT signaling and DNA damage sensing in hepatocellular carcinoma.18 Although it has been reported to promote cell proliferation and metastasis in lung cancer,9 the detailed mechanism remains largely unclear. In this study, we found that CASC9 could regulate HIF-1α expression at both transcriptional and post-translational levels in lung cancer cells, rather than merely stabilizing HIF-1α protein as in nasopharyngeal carcinogenesis.17 This may be due to the different gene expression profiling in different cancers.
HIF-1α is considered as the master transcriptional regulator of cellular and developmental processes in response to hypoxia.19 It is involved in cancer metastasis through many signal pathways.14–16 HIF-1α is also closely implicated in cell proliferation control in many cancers.20–22 In nasopharyngeal carcinoma, CASC9 was reported to interact with HIF-1α and stimulated the expression of its downstream targets.17 Here, we found that knockdown of CASC9 decreased the mRNA and protein mRNA levels of HIF-1α. Specifically, we verified that CASC9 was mainly involved in the proteostasis control of HIF-1α, suggesting that CASC9 may regulate lung cell proliferation and metastasis though affecting the HIF-1α level. Importantly, restoration of HIF-1α by Molidustat, the HIF-stabilizer, majorly recovered cell proliferation, migration and invasion in CASC9-silenced lung cancer cells, further establishing that CASC9 acted through HIF-1α to promote lung cancer progression. On the other hand, induction of HIF-1α significantly increased the expression of CASC9 whereas inhibition of HIF-1α significantly decreased the expression of CASC9. These results showed that HIF-1α can also regulate the expression of CASC9, indicating a positive feedback loop existed between CASC9 and HIF-1α.
Conclusion
In summary, we found that CASC9 forms a positive feedback loop with HIF-1α and promotes cell proliferation and metastasis in lung cancer. Our findings reveal a novel regulatory signaling pathway, namely the CASC9/HIF-1α axis, involved in lung cancer progression. This novel finding provides a potential therapeutic target for lung cancer.
Ethical Approval And Informed Consent
All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration. Informed consent was obtained from all individual participants included in the study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics. 2017;17(2–3):135–143. doi:10.1007/s10142-016-0524-x
3. Xing H, Wang S, Li Q, et al. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed Pharmacother. 2018;105:677–682. doi:10.1016/j.biopha.2018.06.005
4. Xu Y, Deng W, Zhang W. Long non-coding RNA TUG1 protects renal tubular epithelial cells against injury induced by lipopolysaccharide via regulating microRNA-223. Biomed Pharmacother. 2018;104:509–519. doi:10.1016/j.biopha.2018.05.069
5. Pan Z, Mao W, Bao Y, et al. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5(9):2442–2447. doi:10.1002/cam4.2016.5.issue-9
6. Hu X, Li Y, Kong D, et al. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J Cell Physiol. 2019;234(7):10800–10808. doi:10.1002/jcp.v234.7
7. Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25(11):1980–1995. doi:10.1038/s41418-018-0084-9
8. Liu H, Li C, Yang J, et al. Long noncoding RNA CASC9/miR-519d/STAT3 positive feedback loop facilitate the glioma tumourigenesis. J Cell Mol Med. 2018;22(12):6338–6344. doi:10.1111/jcmm.2018.22.issue-12
9. Zhou J, Xiao H, Yang X, et al. Long noncoding RNA CASC9.5 promotes the proliferation and metastasis of lung adenocarcinoma. Sci Rep. 2018;8(1):37. doi:10.1038/s41598-017-18280-3
10. Noh JH, Gorospe M. AKTions by Cytoplasmic lncRNA CASC9 promote hepatocellular carcinoma survival. Hepatology. 2018;68(5):1675–1677. doi:10.1002/hep.v68.5
11. Gao GD, Liu XY, Lin Y, et al. LncRNA CASC9 promotes tumorigenesis by affecting EMT and predicts poor prognosis in esophageal squamous cell cancer. Eur Rev Med Pharmacol Sci. 2018;22(2):422–429. doi:10.26355/eurrev_201801_14191
12. Goossens S, Vandamme N, Van Vlierberghe P, et al. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer. 2017;1868(2):584–591. doi:10.1016/j.bbcan.2017.06.006
13. Liu F, Gu LN, Shan BE, et al. Biomarkers for EMT and MET in breast cancer: an update. Oncol Lett. 2016;12(6):4869–4876. doi:10.3892/ol.2016.5369
14. Lin J, Shi Z, Yu Z, et al. LncRNA HIF1A-AS2 positively affects the progression and EMT formation of colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed Pharmacother. 2018;98:433–439. doi:10.1016/j.biopha.2017.12.058
15. Zhang W, Shi X, Peng Y, et al. HIF-1α promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PLoS One. 2015;10(6):e0129603. doi:10.1371/journal.pone.0129603
16. Zhang L, Huang G, Li X, et al. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor −1α in hepatocellular carcinoma. BMC Cancer. 2013;13:108. doi:10.1186/1471-2407-13-108
17. Su X, Li G, Liu W. The long noncoding RNA cancer susceptibility candidate 9 promotes nasopharyngeal carcinogenesis via stabilizing HIF1α. DNA Cell Biol. 2017;36(5):394–400. doi:10.1089/dna.2016.3615
18. Klingenberg M, Gross M, Goyal A, et al. The long noncoding RNA cancer susceptibility 9 and RNA binding protein heterogeneous nuclear ribonucleoprotein L form a complex and coregulate genes linked to AKT signaling. Hepatology. 2018;68(5):1817–1832. doi:10.1002/hep.v68.5
19. Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12(2):149–162. doi:10.1101/gad.12.2.149
20. Carmeliet P, Dor Y, Herbert JM, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–490. doi:10.1038/28867
21. Mouriaux F, Sanschagrin F, Diorio C, et al. Increased HIF-1α expression correlates with cell proliferation and vascular markers CD31 and VEGF-A in uveal melanoma. Invest Ophthalmol Vis Sci. 2014;55(3):1277–1283. doi:10.1167/iovs.13-13345
22. Zhang Y, Yan J, Wang L, et al. HIF-1α promotes breast cancer cell MCF-7 proliferation and invasion through regulating miR-210. Cancer Biother Radiopharm. 2017;32(8):297–301. doi:10.1089/cbr.2017.2270
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.