Back to Journals » Cancer Management and Research » Volume 13
Long Non-Coding RNA CAR10 Facilitates Non-Small Cell Lung Cancer Cell Migration and Invasion by Modulating the miR-892a/GJB2 Pathway
Authors Li S , Liu Y , Qiu G , Luo Y, Luan L , Xu T, Wang Y, Xia S
Received 18 October 2020
Accepted for publication 8 January 2021
Published 25 February 2021 Volume 2021:13 Pages 1967—1979
DOI https://doi.org/10.2147/CMAR.S287386
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Shanshan Li1 *,* Yize Liu2 *,* Guanzhen Qiu,2 Yinzhou Luo,2 Lan Luan,3 Tiance Xu,4 Yong Wang,2,5 Shuyue Xia1,6
1Respiratory Department, Central Hospital Affiliated to Shenyang Medical College, Shenyang, Liaoning, 110024, People’s Republic of China; 2 4th Department of Orthopedics, Central Hospital Affiliated to Shenyang Medical College, Shenyang, Liaoning, 110024, People’s Republic of China; 3Department of Pathology, Central Hospital Affiliated to Shenyang Medical College, Shenyang, Liaoning, 110024, People’s Republic of China; 4 2nd Department of Neurology, Central Hospital Affiliated to Shenyang Medical College, Shenyang, Liaoning, 110024, People’s Republic of China; 5Central Laboratory, Central Hospital Affiliated to Shenyang Medical College, Shenyang, Liaoning, 110024, People’s Republic of China; 6Dean’s Office, Central Hospital Affiliated to Shenyang Medical College, Shenyang, Liaoning, 110024, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yong Wang
Central Laboratory and 4th Department of Orthopedics, Central Hospital Affiliated to Shenyang Medical College, 5 South Seven West Road, Shenyang, Liaoning, 110024, People’s Republic of China
Email [email protected]
Shuyue Xia
Respiratory Department, Central Hospital Affiliated to Shenyang Medical College, 5 South Seven West Road, Shenyang, Liaoning, 110024, People’s Republic of China
Email [email protected]
Introduction: Non-coding RNAs, including long non-coding (lnc)RNAs and microRNAs (miRs), play crucial roles in numerous malignant tumors, including non-small cell lung cancer (NSCLC).
Methods: The expression levels of chromatin-associated RNA Intergenic 10 (CAR10), gap junction protein beta 2 (GJB2) and miR-892a in NSCLC were evaluated by reanalyzing three Gene Expression Omnibus (GEO) datasets, and performing reverse transcription-quantitative PCR, immunohistochemistry staining and Western blot analysis, accordingly. Functionally, Transwell and Matrigel assays were performed to measure changes in the migration and invasion abilities of the A549 and H1299 cell lines. The targeted binding effects between CAR10 and miR-892a, as well as between miR-892a and GJB2 were confirmed by conducting dual-luciferase reporter and RNA pull-down assays, respectively.
Results: The present study demonstrated that CAR10 was upregulated in patients with NSCLC, which was also associated with a poor prognosis. Functionally, CAR10 was confirmed to be oncogenic and promoted NSCLC cell migration and invasion, using overexpression and knockdown Transwell assays. Furthermore, GJB2 expression was revealed to be upregulated and was positively correlated with CAR10 expression in NSCLC. A further mechanistic study revealed that GJB2 was a downstream target of CAR10, which induced the migration and invasive potential of the A549 and H1299 cell lines. More specifically, miR-892a was found to serve as a bridge between CAR10 and GJB2, via similar miRNA response elements. The RNA pull-down and luciferase assays indicated that miR-892a directly binds both CAR10 and GJB2.
Conclusion: CAR10 promoted NSCLC cell migration and invasion by upregulating GJB2 and sponging miR-892a. These findings illustrated that the CAR10/miR-892a/GJB2 axis may be a novel molecular target for the treatment of NSCLC.
Keywords: chromatin-associated RNA 10, gap junction protein beta 2, microRNA-892a, metastasis, non-small cell lung cancer
Introduction
It has been estimated that ~234,030 new cases of lung and bronchial cancer were diagnosed in the United States in 2018.1 As the most common subtype of lung cancer, non-small-cell lung cancer (NSCLC) accounts for ~85% of all cases of lung cancer.2 Cigarette smoke and air pollution are the primary sources of lung carcinogens, and the aggressive nature of NSCLC leads to an unfavorable prognosis, with a 5-year survival rate of <5%.3 At initial diagnosis, a large proportion of patients with NSCLC also exhibit distant metastases at sites, such as the bone, skin, thyroid and the brain.4,5 Therefore, the identification of novel and effective anti-metastatic molecules for the treatment of NSCLC is of great importance. Long non-coding (lnc)RNAs, which are different RNA transcripts without protein coding ability, have been associated with a diverse number of cellular and biological processes.6,7 Accumulating evidence has suggested that dysregulation of lncRNA expression has been associated with tumorigenesis, metastasis and drug-resistance via numerous mechanisms in multiple different types of malignancy.8,9 Chromatin-associated RNA Intergenic 10 (CAR10) was first revealed to regulate the expression of neighboring genes by modulating the chromatin structure in cis.9 Guo et al demonstrated that CAR10 acted as an oncogene in leiomyomas and that CAR10 knockdown suppressed leiomyoma cell proliferation.10 Furthermore, Ge et al reported that CAR10 was upregulated in lung adenocarcinoma (LUAD), and that CAR10 stabilized zinc finger protein SNAI-1 and −2 mRNA expression levels by sequestering microRNA (miRNA/miR)-30 and miR-203, subsequently promoting the proliferation, migration and invasiveness of LUAD cell lines.11 However, to date, the function of CAR10 in NSCLC has not been fully elucidated.
In the current study, analysis of the Gene Expression Omnibus (GEO) datasets GSE19188, GSE30219 and GSE118370 revealed that CAR10 expression was upregulated in patients with NSCLC, and was also associated with the expression of gap junction protein beta 2 (GJB2). In addition, GJB2 was found to be a downstream gene of CAR10, and CAR10 was also shown to promote GJB2-mediated migration and invasion by sponging miR-892a in NSCLC cell lines.
Materials and Methods
Patients and Tissue Samples
A total of 50 NSCLC and paired adjacent tumor samples were collected during tumorectomy at the Central Hospital Affiliated to Shenyang Medical College (Shenyang, China) between April 2011 and April 2015. All 50 cases were diagnosed with NSCLC according to a definite pathological diagnosis, and were clinically staged based on the Tumor-Node-Metastasis classification of the International Union Against Cancer. Written informed consent was provided from all the patients, and the study was approved by the Institute Research Medical Ethics Committee of the Central Hospital Affiliated to Shenyang Medical College.
Cell Culture
The normal human 16HBE bronchial epithelial cell line, and the NSCLC cell lines, H1975, A549 and H1299 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The 16HBE cell line was cultured in Airway Epithelial Cell Basal Medium (PromoCell, Heidelberg, Germany), while the H1975 cell line was cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.), and the A549 and H1299 cell lines were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.). All culture media were supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA), 100 IU/mL penicillin (Shanghai Baoman Biotechnology Co., Ltd.) and 100 mg/mL streptomycin (Shanghai Baoman Biotechnology Co., Ltd.). All the cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Reverse Transcription-Quantitative (RT-q) PCR
The RT-qPCR procedure was performed as previously described.12 Total RNA was extracted from tissue specimens and cancer cell lines using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA samples were reversed transcribed into cDNA using the RNA PCR kit, and qPCR was performed to detect the mRNA expression levels of CAR10, GJB2 and miR-892a using SYBR® Premix Ex Taq II (all from Takara Biotechnology Co., Ltd.). GAPDH and U6 were used as the internal controls for CAR10/GJB2 and miR-892a expression, respectively. The primers were synthesized by Guangzhou RiboBio Co., Ltd., and the sequences are displayed in Table 1.
 |
Table 1 Primer and Oligonucleotide Sequences Used in the Present Research |
Oligonucleotide and Plasmid Transfection
A total of three specific small interfering (si)RNAs targeting CAR10 and GJB2, as well as a corresponding scramble control siRNA (siSCR), were chemically synthesized by Guangzhou RiboBio Co., Ltd., and transfected into the A549 and H1299 cell lines to knockdown CAR10 or GJB2, respectively. CAR10 and GJB2 overexpression plasmids (oeCAR10 and oeGJB2), and miR-892a mimics and inhibitors were also synthesized by Guangzhou RiboBio Co., Ltd, to overexpress or knockdown miR-892a expression, respectively. All plasmids and oligonucleotides were transfected into the A549 and H1299 cell lines using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions, and as previously reported.13 The sequences of the synthesized oligonucleotides and plasmid primers are listed in Table 1.
Transwell Assay
Transwell and Matrigel assays were performed as previously described.14 Briefly, the A549 and H1299 cell lines were seeded (1x104 cells/well) into the Matrigel-coated (for the invasion assay) or uncoated (for the migration assay) upper chambers of Transwell inserts (BD Biosciences) and placed into 24-well plates. Culture media, with or without 10% FBS, were added to the lower and upper chambers, respectively, and the plates were incubated for 24 h at 37°C. The next day, non-invasive/migratory cells were removed and the cells that had transitioned onto the membrane of the lower chamber were fixed in 3.75% formaldehyde, then stained using crystal violet. An inverted microscope (Olympus Corporation) was used to count the number of cells in five random fields per chamber.
Western Blot Analysis
Total protein was extracted from the A549 and H1299 cell lines using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA), and 10 μg of each sample was quantified using a BCA Protein Assay kit (Beyotime Institute of Biotechnology) according to the manufacture’s protocol. Next, 50 µg protein/lane was separated using 10% SDS-PAGE and transferred onto a polyvinylidene fluoride membrane. After blocking in 5% skimmed milk in TBS-Tween-20 (TBST; 25 mM Tris, pH 7.5, 150 mM NaCl and 0.1% Tween-20) for 1 h at room temperature, anti-GJB2 (Abcam, cat. no. ab65969, dilution rates of 1:250) and anti-β-actin (Abcam, cat. no. ab179467, dilution rates of 1:2500) antibodies were further incubated with the membrane at 4°C overnight. After washing three times with TBST, the membranes were incubated with a secondary antibody (1:2000; cat. no. ab97047; Abcam) at room temperature for 1 h. The protein bands were detected on an X-ray film using an enhanced chemiluminescence detection system.
Immunohistochemistry (IHC) Staining
IHC staining was performed to detect the protein expression levels of GJB2 according to a previously described method.15 NSCLC tissue specimens were fixed in 4% paraformaldehyde, embedded in paraffin and cut into 4-μm thick sections. The sections were subsequently deparaffinized, rehydrated, then incubated in hydrogen peroxide to block endogenous peroxidase/phosphatase activity. Following antigen retrieval, each sample was blocked with 10% goat serum (Bioworld Technology, Inc.), then incubated with a primary antibody (anti-GJB2; 1:200; cat. no. ab65969; Abcam) overnight at 4°C. The samples were then incubated with the secondary antibody (goat anti-rabbit IgG H&L; 1:500; cat. no. ab97047; Abcam) at 37°C for 20 min, prior to the addition of a streptavidin-horseradish peroxidase complex. The sections were then stained with diaminobenzidine tetrahydrochloride (MedChemExpress) and counterstained with hematoxylin (Amresco, LLC). All sections were individually assessed by two experienced pathologists.
Dual-Luciferase Reporter Assay
The luciferase assay was performed as previously described.16 Full-length CAR10 and GJB2 3ʹ-untranslated region (UTR) constructs containing miR-892a binding sites were amplified and cloned into a psiCHECK2 vector (Promega Corporation). The luciferase reporter vector was co-transfected with miR-892a or negative control (NC) mimics into the A549 or H1299 cell lines using Lipofectamine® 3000. The relative luciferase activity was determined using the Dual-Luciferase Reporter Assay System according to the manufacturer’s protocol (Promega Corporation).
RNA Pull-Down Assay
The RNA pull-down assay was performed as previously described.16 Briefly, wild-type and mutant CAR10 transcripts were labeled using a Biotin RNA Labeling Mix (Roche Diagnostics) and T7 RNA polymerase (Roche Diagnostics), treated with RNase-free DNase I (Roche Diagnostics), then purified using the RNeasy Mini kit (Qiagen, Inc.). Subsequently, 1 mg whole-cell lysate from the A549 or H1299 cell lines was incubated with 3 μg purified biotinylated transcripts for 1 h at 25°C. The complexes were isolated using streptavidin agarose beads (Invitrogen; Thermo Fisher Scientific, Inc.). Any RNA present in the pull-down material was detected using RT-qPCR analysis to confirm the enrichment of miR-892a.
Statistical Analysis
All statistical analyses were performed using SPSS v20.0 (IBM Corp) and GraphPad Prism v5.0 (GraphPad Software, Inc.) software. All experiments were repeated in triplicate and the data are presented as the mean ± SD. Associations between CAR10 expression and the clinicopathological features of patients with NSCLC were assessed using either Pearson’s χ2 or Fisher’s exact tests. Survival analysis was performed using the Log rank test in GraphPad Prism v5.0. A Student’s t-test was used when the variance between groups was equal, and the Wilcoxon signed rank test was used when the variance between groups was unequal. P<0.05, from a two-tailed test, was considered to indicate a statistically significant difference.
Results
CAR10 Expression Levels are Upregulated and Associated with a Poor Prognosis in Patients with NSCLC
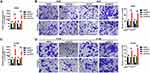
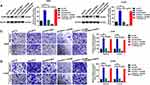
Firstly, CAR10 mRNA expression in the collected NSCLC and paired adjacent tumor tissue samples was quantified using RT-qPCR. As indicated in Figure 1A and B, CAR10 was upregulated in the majority of the NSCLC tissue samples (44/50; 88.00%; P<0.0001; Figure 1B). CAR10 expression was also analyzed using the GEO datasets, and as previously reported by Li et al,11 was markedly upregulated in the GSE19188, GSE30219 and GSE118370 datasets (Figure 1C–E). In addition, the association between CAR10 expression and lymph node metastasis was assessed; the expression of CAR10 was notably higher in patients with NSCLC and lymph node metastasis (N1 and N2) compared with that in patients without lymph node metastasis (N0) (Figure 1F). To evaluate the clinical role of CAR10 in NSCLC, the 50 collected samples were divided into CAR10-high and CAR10-low groups (n=25 each) according to the median expression value, and the association between CAR10 expression and clinicopathological features was evaluated. As illustrated in Figure 1G and Table 2, high CAR10 expression was significantly associated with a shorter survival time (determined using Kaplan-Meier analysis; P=0.014), a higher pathological grade (P=0.000) and more frequent lymph node metastasis (P=0.018). Finally, CAR10 expression was determined in a normal human bronchial epithelial cell line (16HBE) and four NSCLC cell lines (A549, H1975, SPCA1 and H1299). CAR10 expression was significantly upregulated in the four NSCLC cell lines compared with that in the 16HBE cells (P<0.001; Figure 1H).
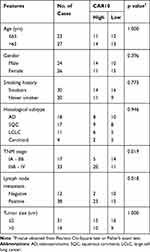 |
Table 2 Association of CAR10 Expression with Clinicopathological Features of NSCLC |
CAR10 Promotes Migration and Invasion in the A549 and H1299 Cell Lines
The role of CAR10 in NSCLC cell migration and invasion was subsequently investigated. Of the three specific CAR10 siRNAs, the most efficient siRNA (siCAR10-2) was selected for use in the RNA interference experiments (Supplementary Figure 1A and B). Furthermore, CAR10 overexpression plasmids (oeCAR10) were synthesized and transfected into the A549 and H1299 cell lines, and RT-qPCR was used to confirm the mRNA expression levels of CAR10. As presented in Figure 2A and C, CAR10 expression was down- and upregulated following transfection with CAR10 siRNAs and oeCAR10, respectively. Furthermore, a subsequent Transwell assay was performed to determine the changes in metastatic ability in A549 and H1299 cell lines. As represented in Figure 2B and D, up- and downregulation of CAR10 promoted and suppressed migration and invasion, respectively, in the A549 and H1299 cell lines (P<0.01).
GJB2 is Upregulated and Positively Correlated with CAR10 in NSCLC
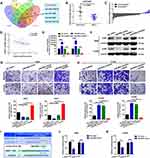
To further investigate the potential downstream targets of CAR10 in NSCLC, the differentially expressed genes (DEGs) in GSE19188, GSE30219 and GSE118370 were analyzed. The top 100 upregulated DEGs are displayed in Figure 3A. Ultimately, 8 genes including GJB2 were filtered out, as they overlapped in the 3 profiling arrays (Supplementary Tables I–III). GJB2 was selected for further investigation, as it was significantly associated with poor prognosis in patients with NSCLC, which was determined using Kaplan-Meier Plotter analysis software (Figure 3B).17 GJB2 was markedly upregulated in all three datasets, GSE19188, GSE30219 and GSE118370 (P<0.0001, P=0.0106 and P=0.0029, respectively) (Figure 3C–E), and gradually increased with advanced staging of NSCLC (Figure 3F). Furthermore, GJB2 was found to be upregulated in the NSCLC cell lines (Figure 3G). Finally, the Pearson’s correlation coefficient test illustrated that GJB2 expression was positively correlated with that of CAR10, according to the data from the aforementioned profiling arrays (P<0.0001, P<0.0001 and P=0.0359) (Figure 3H–J).
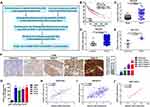 |
Figure 3 GJB2 is upregulated and positively associated with CAR10 expression in NSCLC. (A) Diagram of screening the DEGs in the GEO datasets, GSE19188, GSE30219 and GSE118370 using the GEO2R online software. (B) Prognostic value of GJB2 in 1145 patients with NSCLC was determined using the Kaplan-Meier Plotter tool (https://kmplot.com/analysis/). GJB2 expression in NSCLC and normal lung tissues or ANTs was analyzed in (C) GSE19188, (D) GSE30219 and (E) GSE118370. (F) Expression of GJB2 in patients at different stages of NSCLC was detected using immunohistochemical analysis. *P=0.0106, **P=0.0029, ****P<0.0001. Magnification, x400. Scale bar, 50 μm. (G) GJB2 expression in a normal human bronchial epithelial cell line (16HBE) and in four NSCLC cell lines (A549, H1975, SPCA1 and H1299) was determined using reverse transcription-quantitative PCR. ***P<0.001. Correlation between CAR10 and GJB2 expression in the (H) GSE19188, (I) GSE32219 and (J) GSE118370 GEO datasets. (J) was assessed using Pearson’s correlation analysis. The data are presented as the mean ± SD from three independent experiments. Abbreviations: GJB2, gap junction protein beta 2; CAR10, chromatin-associated RNA Intergenic 10; NSCLC, non-small cell lung cancer; DEG, differentially expressed gene; GEO, Gene Expression Omnibus; ANT, adjacent normal tissue. |
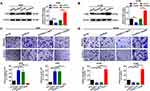
CAR10 Promotes Migration and Invasion by Upregulating GJB2 in the A549 and H1299 Cells
The aforementioned findings indicated that GJB2 was upregulated in NSCLC, and positively correlated with the expression of CAR10. Therefore, the role of GJB2 in CAR10-induced migration and invasion was investigated further. Up- and downregulation of CAR10 was positively regulated by GJB2 protein expression in the A549 and H1299 cells (Figure 4A and B). In addition, three GJB2 siRNAs were constructed and the most efficient was selected for the following GJB2-RNA interference experiments (Supplementary Figure 2A and B). A Transwell assay was also performed, and as displayed in Figure 4C and D, the overexpression of CAR10 promoted A549 and H1299 cell migration and invasion, and this effect was abolished by knocking down GJB2 (co-transfection with oeCAR10 and siGJB2). Conversely, CAR10 knockdown suppressed the migratory and invasive abilities of the A549 and H1299 cell lines, which was reversed by GJB2 overexpression. In brief, these findings indicated that CAR10 promoted migration and invasion in the A549 and H1299 cells by upregulating GJB2.
GJB2 is a Direct Target of miR-892a
A number of studies have recently demonstrated that lncRNAs function as miRNA sponges to regulate their downstream targets. In the current study, several miRNAs were also found to be involved in the CAR10/GJB2 axis-mediated migration and invasion of the A549 and H1299 cells. Using online bioinformatics tools RegRNA 2.0, miRWalk, miRDB and TargetScan,18–21 five miRNAs that may interact with GJB2 or CAR10 were found to overlap in all four of the databases evaluated (Figure 5A). miR-892a was selected for further investigation, as it was downregulated in a miRNA profiling array of NSCLC (GSE19945) (Figure 5B). miR-892a was also frequently downregulated in the 50 collected NSCLC tissue samples (39/50; 78.00%; Figure 5C and Supplementary Figure 3A) and in NSCLC cell lines (Supplementary Figure 3B), and was found to be negatively correlated with the expression of GJB2 (Figure 5D). Furthermore, the up- and downregulation of miR-892a was negatively regulated by GJB2 protein expression (Figure 5E and F), and the upregulation of miR-892a was functionally demonstrated to suppress A549 and H1299 cell migration and invasion; this suppressive effect was reversed by upregulating GJB2 expression (co-transfection of miR-892a mimics and oeGJB2) (Figure 5G). By contrast, downregulation of miR-892a (transfection with an miR-892a inhibitor) enhanced the migratory and invasive abilities of the A549 and H1299 cell lines, which were attenuated by GJB2 inhibition (co-transfection with miR-892a inhibitor and siGJB2) (Figure 5H). A wild-type reporter plasmid (GJB2-luc-WT) containing miR-892a binding sites, and a mutant reporter plasmid (GJB2-luc-MUT) with mutated miR-892a binding sequences, were constructed to confirm the targeted binding effect between GJB2 and miR-892a (Figure 5I). A luciferase reporter assay was then performed. As showed in Figure 5J and K, co-transfection of GJB2-luc-WT and miR-892a mimics significantly decreased the luminescence of the A549 and H1299 cell lines. When the miR-892a seeding sequences in GJB2 were mutated, luminescence was restored. Collectively, these findings suggested that miR-892a is an upstream regulator of GJB2, which itself regulates A549 and H1299 cell migration and invasion by suppressing GJB2.
CAR10 Upregulates GJB2 Partially by Sequestering miR-892a
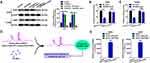
As aforementioned, CAR10 was indicated to promote NSCLC cell migration and invasion by upregulating GJB2, and GJB2 was revealed to be a direct target of miR-892a. Therefore, the association between CAR10, miR-892a and GJB2 was investigated further. lncRNAs are known to regulate downstream mRNAs by sponging miRNAs (the competitive endogenous RNA theory), which was first proposed by Leonardo Salmena.22 In the present study, CAR10 was hypothesized to regulate GJB2 by miR-892a sequestration, and was found to upregulate GJB2 at the protein level (Figure 4A). Notably, the facilitative effect of CAR10 was significantly abrogated by miR-892a overexpression. Furthermore, when the CAR10 miR-892a binding sites were mutated, the promotive effect on GJB2 was abolished (Figure 6A) (P<0.01). A luciferase assay was also performed to detect the targeted binding effect between CAR10 and miR-892a. As shown in Figure 6B and C, co-transfection of a WT CAR10 reporter plasmid (CAR10-luc-WT, containing WT miR-892a binding sequences) and miR-892a mimics resulted in a marked decrease in luminescence. When the predicted seeding sequences were mutated (co-transfection of CAR10-luc-MUT and miR-892a mimics), the luminescence was restored (P<0.01). These phenomena indicated that CAR10 could directly bind with miR-892a. Furthermore, cell lysates from the A549 and H1299 cell lines were incubated with biotin-labeled CAR10 (biotin-CAR10-WT and biotin-CAR10-MUT, containing wild-type and mutant miR-892a binding sequences, respectively), then RT-qPCR was performed following RNA pull-down (to evaluate the retrieved miR-892a). Figure 6D and E indicated that miR-892a was significantly enriched in the biotin-CAR10-WT group compared with that in the biotin-CAR10-MUT group (P<0.0001).
CAR10 Promotes Migration and Invasion by Regulating the miR-892a/GJB2 Axis
Finally, CAR10 overexpression (oeCAR10) and mutant plasmids with mutated miR-892a binding sequences (oeCAR10-MUT) were transfected into the A549 and H1299 cells. GJB2 expression was evaluated using Western blot analysis (Figure 7A and B). A Transwell assay was then performed to determine the effect of the CAR10/miR-892a/GJB2 axis on NSCLC cell migration and invasion. As illustrated in Figure 7C and D, compared with that in the vector control group, oeCAR10, but not oeCAR10-MUT, promoted migration and invasion in the A549 and H1299 cell lines; the facilitative effect of oeCAR10 was reversed by GJB2 knockdown (co-transfection with oeCAR10 and siGJB2). In addition, further suppression of miR-892a abolished the inhibitory effect of siGJB2 in the NSCLC cells. Taken together, these results illustrated that CAR10 promoted migration and invasion by regulating the miR-892a/GJB2 axis.
Discussion
Accumulating evidence indicates that ncRNAs, including lncRNAs and miRNAs, contribute to carcinogenesis and cancer progression. lncRNAs are known to regulate gene expression at local transcriptional sites (in-cis) and at distinct independent loci (in-trans).7 CAR10 was first reported by Kanduri et al in a deep sequencing study of CARs from human fibroblasts.9 They also revealed that CAR10 is positioned between two protein-coding RNAs, fibronectin and ADAM metallopeptidase domain 12. Li et al reported that CAR10 promoted epithelial-to-mesenchymal transition (EMT) by directly binding to miR-30 and miR-203, and subsequently regulating the expression level of SNAI-1 and −2 in lung adenocarcinoma.11 The present study also focused on the expression of CAR10 in three NSCLC-associated expression profiling arrays from GEO datasets, which revealed that CAR10 was significantly upregulated in NSCLC. In addition, high CAR10 expression levels were associated with lymph node metastasis and poor prognosis; thus, the role of CAR10 in NSCLC cell metastatic ability was evaluated. Using overexpression and knockdown assays, CAR10 was demonstrated to serve as an oncogene and promote NSCLC cell metastasis.
To determine the potential downstream target of CAR10, the top 100 upregulated genes in the GSE19188, GSE30219 and GSE118370 datasets were identified. GJB2 was selected, as it overlapped in all three datasets, and high GJB2 expression was associated with poor prognosis in 1145 patients with NSCLC. GJB2 (also known as connexin 26), is located on chromosome 13q12.11 and contains three exons. GJB2 is considered to be an oncogene and has been associated with tumor growth, EMT and lymph node metastasis in multiple types of cancer.23–26 The results of the present study indicated that GJB2 expression was positively correlated with that of CAR10, and CAR10 was revealed to positively regulate GJB2 protein expression. Furthermore, CAR10 promoted NSCLC cell migration and invasion by upregulating GJB2 expression, which collectively suggested that GJB2 is a downstream target in CAR10-induced NSCLC cell migration and invasion.
A high number of studies have demonstrated that lncRNAs function as miRNA sponges to regulate the expression of mRNA.13,16,27 lncRNAs serve as miRNA decoys via their miRNA response elements, and protect the corresponding downstream mRNAs from degradation.22 A previous study demonstrated that CAR10 was primarily located in the cytoplasm and served as miR-30 and miR-203 sponges to promote SNAI-1 and −2 expression.11 CAR10 was therefore considered to regulate GJB2 using a similar mechanism. Using online bioinformatics tools, the present study revealed that miR-892a was the key molecule connecting CAR10 and GJB2. Analyzing the GEO GSE19945 dataset revealed that miR-892a was downregulated in NSCLC, and subsequent RT-qPCR detection in patient tissues also demonstrated similar results. miRNAs are known to directly bind and regulate their target genes. Using luciferase reporter and RNA pull-down assays, the present study demonstrated that both GJB2 and CAR10 were targets of miR-892a, and that miR-892a suppressed NSCLC cell metastasis by directly binding to the GJB2 3ʹUTR. Transwell assays also revealed that CAR10 promoted NSCLC cell migration and invasion via GJB2 upregulation by sponging miR-892a.
Conclusion
Cancer metastasis is a complex biological process, that is facilitated by numerous molecules and signaling pathways. The results of the present study indicated that CAR10 served as an oncogene by promoting the migration and invasion abilities of the NSCLC cells. CAR10 was also shown to enhance the expression of GJB2 by acting as a decoy for miR-892a. As a regulator of tumor metastasis, CAR10 could have significant potential as a diagnostic and prognostic marker, and as a therapeutic target in NSCLC. Therefore, the CAR10/miR-892a/GJB2 axis may present a novel target for the molecular treatment of NSCLC.
Ethics Approval and Consent to Participate
The current research was conducted in in accordance with the Declaration of Helsinki and ethics approval was obtained from the Institute Research Medical Ethics Committee of the Central Hospital Affiliated to Shenyang Medical College. Written informed consent was provided by each participant.
Acknowledgments
The present study was supported by grants from the Key R&D Program of Liaoning Province (grants no. 2018225014), the National Natural Science Foundation of China (grant no. 81972522 and 81502333), Youth Talent Support Program of Liaoning Province (grants No. XLYC1907011), Technological innovation fund of Shenyang Technology Division (No. RC190008 and 19-112-4-023), Shenyang Science and Technology Plan Project (No. 17-230-9-05) and the SMC Students’ scientific research projects (grant no. Y20190520).
Disclosure
The authors declare that they have no competing interests.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi:10.3322/caac.21442
2. Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–719. doi:10.1016/S0140-6736(13)61502-0
3. Velcheti V, Schalper KA, Carvajal DE, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Investig. 2014;94:107–116. doi:10.1038/labinvest.2013.130
4. Chen S, Zhang Z, Zhang X, et al. TCM therapies combined with chemotherapy for preventing recurrence and metastasis in postoperative II to IIIA NSCLC: A protocol for a systematic review and meta-analysis. Medicine. 2019;98:e14724. doi:10.1097/MD.0000000000014724
5. Tan PS, Bilger M, de Lima Lopes G, Acharyya S, Haaland B. Meta-analysis of first-line therapies with maintenance regimens for advanced non-small-cell lung cancer (NSCLC) in molecularly and clinically selected populations. Cancer Med. 2017;6:1847–1860.
6. Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi:10.1016/j.tibs.2016.07.003
7. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi:10.1016/j.cell.2018.01.011
8. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi:10.1038/nm.3981
9. Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi:10.1101/gr.103473.109
10. Guo H, Zhang X, Dong R, et al. Integrated analysis of long noncoding RNAs and mRNAs reveals their potential roles in the pathogenesis of uterine leiomyomas. Oncotarget. 2014;5:8625–8636. doi:10.18632/oncotarget.2349
11. Ge X, Li GY, Jiang L, et al. Long noncoding RNA CAR10 promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis. Oncogene. 2019;38:3061–3076. doi:10.1038/s41388-018-0645-x
12. Wang Y, Zhang Y, Yang T, et al. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8:59417–59434. doi:10.18632/oncotarget.19727
13. Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi:10.1186/s12943-018-0837-6
14. Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–867. doi:10.1111/cas.13201
15. Wang Y, Yang T, Liu Y, et al. Decrease of miR-195 promotes chondrocytes proliferation and maintenance of chondrogenic phenotype via targeting FGF-18 pathway. Int J Mol Sci. 2017;18.
16. Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. doi:10.1038/s41467-017-00304-1
17. Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi:10.1038/s41598-018-27521-y
18. Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinform. 2013;14(Suppl 2):S4. doi:10.1186/1471-2105-14-S2-S4
19. Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13:e0206239. doi:10.1371/journal.pone.0206239
20. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi:10.1093/nar/gku1104
21. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4. doi:10.7554/eLife.05005
22. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi:10.1016/j.cell.2011.07.014
23. Ezumi K, Yamamoto H, Murata K, et al. Aberrant expression of connexin 26 is associated with lung metastasis of colorectal cancer. Clin Cancer Res. 2008;14:677–684. doi:10.1158/1078-0432.CCR-07-1184
24. Inose T, Kato H, Kimura H, et al. Correlation between connexin 26 expression and poor prognosis of esophageal squamous cell carcinoma. Ann Surg Oncol. 2009;16:1704–1710. doi:10.1245/s10434-009-0443-3
25. Naoi Y, Miyoshi Y, Taguchi T, et al. Connexin26 expression is associated with aggressive phenotype in human papillary and follicular thyroid cancers. Cancer Lett. 2008;262:248–256. doi:10.1016/j.canlet.2007.12.008
26. Wu JI, Wang LH. Emerging roles of gap junction proteins connexins in cancer metastasis, chemoresistance and clinical application. J Biomed Sci. 2019;26:8. doi:10.1186/s12929-019-0497-x
27. Wang Y, Xu Z, Jiang J, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi:10.1016/j.devcel.2013.03.002
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.