Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA C1QTNF1 Antisense RNA 1 Upregulates Hexokinase 2 by Sponging microRNA-484 to Promote the Malignancy of Colorectal Cancer
Authors Jin S, Liu Y, Wang W, Li Z
Received 29 July 2020
Accepted for publication 3 November 2020
Published 24 November 2020 Volume 2020:12 Pages 12053—12066
DOI https://doi.org/10.2147/CMAR.S262096
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Seema Singh
This paper has been retracted.
Shifeng Jin, 1 Yi Liu, 1 Wenjing Wang, 1 Ze Li 2
1Department of Anorectal, The Affiliated Hospital of Changchun University of Chinese Medicine, Changchun, Jilin 133021, People’s Republic of China; 2Department of Colorectal and Stomach Cancer Surgery-1, Jilin Cancer Hospital, Changchun, Jilin 130000, People’s Republic of China
Correspondence: Ze Li
Department of Colorectal and Stomach Cancer Surgery-1, Jilin Cancer Hospital, 1018 Huguang Road, Changchun, Jilin 130000, People’s Republic of China
Email [email protected]
Purpose: The long noncoding RNA C1QTNF1 antisense RNA 1 (C1QTNF1-AS1) contributes to hepatocellular carcinoma development. However, its expression and roles in colorectal cancer (CRC) have not been fully explored. Therefore, this study determined the expression and roles of C1QTNF1-AS1 in CRC and elucidated its detailed mechanism of action.
Methods: C1QTNF1-AS1 expression in CRC tissues and cell lines was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). We used Cell Counting Kit-8, flow cytometry, cell migration and invasion assays, and a xenograft tumor model to test the effects of C1QTNF1-AS1 on CRC malignancy. The associations among C1QTNF1-AS1, microRNA-484 (miR-484), and hexokinase 2 (HK2) were explored using luciferase reporter assay, RNA immunoprecipitation, RT–qPCR, and Western blotting.
Results: C1QTNF1-AS1 was overexpressed in CRC and related to poor prognosis. C1QTNF1-AS1 interference inhibited CRC cell proliferation, migration, and invasion but induced apoptosis. Furthermore, C1QTNF1-AS1 deficiency impaired tumor growth in vivo. Mechanistically, C1QTNF1-AS1 adsorbed miR-484, thereby increasing the expression of its target HK2. Rescue experiments revealed that the effects of C1QTNF1-AS1 deficiency in CRC cells were reversed by inhibiting miR-484 or upregulating HK2.
Conclusion: C1QTNF1-AS1 drives CRC progression by sponging miR-484 and consequently upregulating HK2. The C1QTNF1-AS1/miR-484/HK2 pathway may serve as a diagnostic and therapeutic target for CRC.
Keywords: competitive endogenous RNA, therapeutic target, hexokinase 2
Introduction
Colorectal cancer (CRC) is the third most frequent human cancer and the second leading cause of cancer-associated deaths globally.1 Every year, ~1.2 million novel cases are diagnosed and 860,000 mortalities occur worldwide.2 Surgical excision, chemoradiotherapy, and immunotherapy are the currently available effective therapeutic techniques for early stages of CRC.3 Despite substantial improvements in diagnostic methods and therapies, the treatment outcomes of patients with advanced-stage CRC remain unsatisfactory.4,5 Approximately 50–60% of CRC patients will experience recurrence and metastasis even after surgery, and it is now known that these are the major causes of death among patients with CRC.6,7 Increased tumor suppressor gene inactivation and oncogene activation drive CRC pathogenesis by deregulating important signaling pathways; however, the detailed mechanisms have not been clearly elucidated.8–10 Therefore, it is an essential and urgent need to acquire an in-depth comprehension of the molecular mechanisms associated with CRC oncogenesis and progression to develop promising new targets for CRC diagnosis, prognosis, and management.
Long noncoding RNAs (lncRNAs) comprise a family of transcripts longer than 200 nucleotides in length.11 They lack protein-coding ability and are expressed in a highly context-specific pattern.12 lncRNAs play key roles in normal physiological processes and pathological behaviors by controlling gene expression at the transcriptional and posttranscriptional levels.13 In recent years, extensive studies have revealed that alterations in lncRNA expression are closely related to diverse human diseases, particularly cancers.14 Increasing evidence has demonstrated that a number of lncRNAs are dysregulated in CRC, contributing to CRC genesis and progression.15–17 LncRNAs can execute cancer-inhibiting or cancer-promoting activities during CRC oncogenesis and are directly involved in several malignant processes.18,19
MicroRNAs (MiRNAs) are a family of short noncoding RNA molecules with a length of approximately 17–24 nucleotides.20 MiRNAs have been shown to actively regulate gene expression by base complementary pairing with the 3′-untranslated regions (3′-UTRs) of their target genes, resulting in mRNA degradation or translational suppression.21 Changes in miRNA expression in CRC have been widely reported, and their abnormal expression has been notably correlated with CRC tumorigenesis.22–24 Recently, the competitive endogenous RNA (ceRNA) hypothesis was proposed and is now widely accepted.25,26 lncRNAs are a type of ceRNA that work as molecular sponges for miRNAs, regulating target mRNAs at the posttranscriptional level.27 Accordingly, a comprehensive understanding of the roles of lncRNAs and miRNAs in CRC may offer novel insights into CRC pathogenesis and thus improve the therapeutic options.
The lncRNA C1QTNF1 antisense RNA 1 (C1QTNF1-AS1) has been reported to contribute to the tumorigenesis of hepatocellular carcinoma.28,29 However, its expression and detailed functions in CRC are not well understood. This study aimed to determine C1QTNF1-AS1expression in CRC, investigate the important roles played by C1QTNF1-AS1 in CRC cells, and elucidate the underlying molecular events.
Materials and Methods
Patients and Clinical Specimens
CRC tissues and corresponding adjacent normal tissues were collected from 71 patients admitted to the Jilin Cancer Hospital. No patients had been previously treated with preoperative anticancer therapies or presented with cancers other than CRC. The present study was approved by the Ethics Committee of Jilin Cancer Hospital and conducted in accordance with the Declaration of Helsinki. Moreover, written informed consent forms were collected from all participants. Following tissue excision, all tissues were immediately frozen and stored in liquid nitrogen until further use.
Cell Lines
Four human CRC cell lines, namely HCT116, HT29, SW480, and SW620, were obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). HCT116 and HT29 cell lines were grown in McCOY’s 5A medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific Inc.), 100 U/mL penicillin, and 100 mg/mL streptomycin (Gibco; Thermo Fisher Scientific Inc.). L-15 medium (Gibco; Thermo Fisher Scientific Inc.) supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin was used to culture the other two CRC cell lines.
A normal human colon epithelium cell line (FHC) was obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in DMEM:F12 medium (Gibco; Thermo Fisher Scientific Inc.) with 25 mM HEPES, 10 ng/mL cholera toxin, 0.005 mg/mL insulin, 0.005 mg/mL transferrin, 100 ng/mL hydrocortisone, 20 ng/mL human recombinant epidermal growth factor, and 10% FBS. The culture conditions for all cells were 95% humidity, 5% CO2, and 37°C.
Small Interfering RNA (siRNA), Vector, miRNA Mimic, and miRNA Inhibitor Transfections
The siRNAs used to silence C1QTNF1-AS1 expression (si-C1QTNF1-AS1) and negative control siRNA were designed and produced by RiboBio (Guangzhou, China). MiR-484 mimic, negative control miRNA mimic (miR-NC), miR-484 inhibitor, and negative control (NC) inhibitor were obtained from GenePharma Co., Ltd. (Shanghai, China). The pcDNA3.1 vectors overexpressing HK2 (pcDNA3.1-HK2) were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). An empty pcDNA3.1 vector was used as the negative control for pcDNA3.1-HK2. Logarithmic growth phase CRC cells were inoculated in 6-well plates, and cell transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) after the cell density reached 80% confluence.
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was isolated using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the supplier’s protocol. For detecting C1QTNF1-AS1 and HK2 mRNA expression, total RNA was reverse transcribed into cDNA using a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). Using the SYBR Premix Ex Taq™ Kit (Takara Biotechnology Co., Ltd.), qPCR was conducted on an ABI 7900 Real-Time PCR system (Applied Biosystems, Foster City, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control, and C1QTNF1-AS1 and HK2 expression was normalized to GAPDH expression.
To measure miR-484 expression, reverse transcription was performed using a miScript Reverse Transcription kit (Qiagen GmbH, Hilden, Germany); subsequently, the obtained cDNA was used as a template for PCR amplification using a miScript SYBR Green PCR kit (Qiagen GmbH). MiR-484 expression was normalized to U6 small nuclear RNA expression. The 2−ΔΔCt method was employed to analyze relative gene expression.
Subcellular Fractionation
The nuclear and cytosolic fractions of CRC cell suspensions were isolated using a Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA). Both fractions were subjected to RT–qPCR to determine the localization of C1QTNF1-AS1 expression in CRC cells. GAPDH and U6 were used as cytoplasmic and nuclear controls, respectively.30–32
Cell Counting Kit-8 (CCK-8) Assay
Twenty-four hours after transfection, the cells were harvested and seeded into 96-well plates at a density of 300 cells/well. After the addition of 10 μL CCK8 solution (Dojindo Laboratories Co., Ltd., Kumamoto, Japan), the cells were cultured in an incubator at 37°C with 5% CO2 for an additional 2 h, followed by detecting the absorbance at 450 nm using a Multiskan Ex microtiter plate reader (Labsystems, Helsinki, Finland). The CCK-8 assay was conducted at 0, 24, 48, and 72 h after cell inoculation and proliferation curves were generated.
Flow Cytometry
The transfected cells were collected, rinsed with pre-cooled phosphate buffer solution (Gibco; Thermo Fisher Scientific Inc.), and subjected to apoptotic analysis using an Annexin V–Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (BioLegend, San Diego, CA, USA). After centrifugation, the supernatant was discarded, and the resultant cells were resuspended in 100 μL of flow cytometry binding buffer. Cells were double-stained with 5 μL of Annexin V-FITC and 5 μL of propidium iodide, and the proportion of apoptotic cells was measured using a flow cytometer (FACScan™, BD Biosciences, Franklin Lakes, NJ, USA).
Cell Migration and Invasion Assays
To determine cell migration, transfected cells after 48 h of incubation were mixed with FBS-free culture medium to obtain a single-cell suspension. The upper chambers of 24-well Transwell plates (BD Biosciences) were loaded with 100 μL of cell suspension containing 5 × 104 cells. A volume of 600 μL culture medium supplemented with 20% FBS was added into the lower chambers. After 24 h of incubation at 37°C, the migrated cells were fixed in 4% paraformaldehyde and stained using 0.5% crystal violet. The stained cells were then photographed and counted under a light microscope (Olympus Corporation, Tokyo, Japan) at a magnification of ×200. To examine cell invasion, the Transwell plates were coated with Matrigel (BD Biosciences) and incubated at 37°C for 3 h in an incubator. The same experimental procedures were followed as described above for the migration assay.
Xenograft Tumor Model
The short hairpin RNA (shRNA) targeting C1QTNF1-AS1 (sh-C1QTNF1-AS1) and negative control shRNA (sh-NC) were produced by RiboBio and inserted into the lentivirus vector pLKO.1 vector. The generated vectors, namely pLKO.1-sh-C1QTNF1-AS1 and pLKO.1-sh-NC, alongside psPAX2 and pMD2.G were transduced into 293T cells (Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences). After 48 h, the lentiviruses expressing either sh-C1QTNF1-AS1 or sh-NC were introduced into SW480 cells. Cells stably expressing sh-C1QTNF1-AS1 or sh-NC were selected with puromycin (5 μg/mL; Sigma-Aldrich; Merck KGaA).
BALB/c nude mice (male, aged 4–6 weeks) were purchased from Shanghai SLAC Laboratory Animal, Co., Ltd. (Shanghai, China) and subcutaneously injected with 1 × 106 SW480 cells stably expressing sh-C1QTNF1-AS1 or sh-NC. The tumor size was recorded every 7 days, and tumor volume was calculated using the following formula: Volume = 0.5 × width2 × length. At 28 days post-injection, all mice were euthanized through cervical vertebrae luxation, and the resulting tumor xenografts were resected, weighed, and stored for further use. The animal experimental protocol was approved by the Ethics Committee of Jilin Cancer Hospital and complied with the Animal Protection Law of the People’s Republic of China-2009 for experimental animals.
Bioinformatic Analyses
miRDB (http://mirdb.org/) was used to identify target miRNAs of C1QTNF1-AS1. The putative targets of miR-484 were predicted using three bioinformatics tools: miRDB, TargetScan (http://www.targetscan.org/), and starBase 3.0 (http://starbase.sysu.edu.cn/). The expression of miR-484 in CRC was analyzed using YM500v3 (http://120.110.158.132:8787/ym500v3/).
RNA Immunoprecipitation (RIP) Assay
RIP assays were conducted using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). CRC cells were harvested via centrifugation and treated with RIP buffer. The resultant cell lysate was incubated overnight at 4°C with magnetic beads conjugated to human anti-Argonaute 2 (Ago2) or control IgG antibody (Millipore). Proteinase K was used to digest protein from immunoprecipitates. Finally, RT–qPCR was performed to quantify C1QTNF1-AS1 and miR-484 enrichment in the immunoprecipitated RNA.
Luciferase Reporter Assay
The fragments of C1QTNF1-AS1 (420 bp) harboring the wild-type (wt) target sequence of miR-484 and mutant (mut) C1QTNF1-AS1 fragments were amplified using RT–qPCR and inserted into the pmirGLO dual-luciferase reporter vector (Promega Corporation, Madison, WI, USA). The resultant luciferase reporter vectors were termed C1QTNF1-AS1-wt and C1QTNF1-AS1-mut, respectively. The same experimental procedures were used to construct HK2-wt and HK2-mut luciferase reporter vectors (420 bp). The primers were designed as follows: C1QTNF1-AS1-wt, 5′-CCGCTCGAGAGAGAACTAGAGGCTGCAGCG−3′ (forward) and 5′-GCTCTAGACCAGTGGCCAGGGAGGCTGGAT-3′ (reverse); C1QTNF1-AS1-mut, 5′-CCGCTCGAG AGAGAACTAGAGGCTGCAGCG−3′ (forward) and 5′-GCTCTAGACCAGTGGCCAGGGAGGCTGGAT-3′ (reverse); HK2-wt, 5′-CCGCTCGAGGTAATCCCCCTTGCCAAATTC−3′ (forward) and 5′-GCTCTAGAACATGTGCGCCCCGCTGAGAT−3′ (reverse); and HK2-mut, 5′-CCGCTCGAGGTAATCCCCCTTGCCAAATTC−3′ (forward) and 5′-GCTCTAGAACATGTGCGCCCCGCTGAGAT−3′ (reverse).
CRC cells were seeded into 24-well plates with a density of 1.5 × 105 cells per well. The wt (1.6 μg) or mut (1.6 μg) reporter vectors alongside miR-484 mimic (40 pmol) or miR-NC (40 pmol) were cotransfected into CRC cells using Lipofectamine® 2000 (2.0 μL). We quantified the luciferase activity using a dual-luciferase reporter assay system (Promega Corporation) and normalized to Renilla activity. Luciferase and renilla activities were quantified employing GloMax luminometer (Promega Corporation).
Western Blotting
Cultured cells were lysed using the RIPA Lysis Buffer (Beyotime Institute of Biotechnology; Shanghai, China), and total protein concentration was detected using bicinchoninic acid assay (Beyotime Institute of Biotechnology). SDS–PAGE (10%) was used to resolve equal amounts of protein. The membranes were blocked with 5% milk in TBS–Tween-20 buffer (TBST) for 2 h at room temperature prior to overnight incubation at 4°C with primary antibodies against HK2 (cat. No. ab209847; Abcam, Cambridge, UK) or GAPDH (cat. No. ab128915; Abcam). Subsequently, the membranes were incubated with the goat anti-rabbit horseradish peroxidase-conjugated secondary antibodies (cat. No. ab150077; Abcam) at room temperature for 2 h. After 3 rinses with TBST, the protein bands were visualized using the PierceTM ECL Western blotting substrate (ThermoFisher Scientific). GAPDH was considered the loading control for normalization.
Microsatellite Instability (MSI) Analysis
The MSI status of tumors was tested using a multiplex PCR method. A total of four mononucleotide repeat markers, including BAT26, NR21, NR27 and CAT25, were analyzed. Tumors presenting MSI in ≥1 mononucleotide repeat marker were classified as MSI phenotype, whereas those without MSI were classified as non-MSI phenotype.
Statistical Analysis
All data were presented as mean ± standard deviation from at least three independent replicates. The correlations of C1QTNF1-AS1 expression with clinical variables of patients with CRC were analyzed using chi-squared test. Survival curves were obtained using the Kaplan–Meier method, and the Log rank test was applied to determine the differences between curves. Pearson’s correlation coefficient was used to evaluate the expression relationship between the two genes in CRC tissues. Differences between two groups were examined using Student’s t-test. One-way ANOVA followed by Tukey’s post hoc test was used for comparisons among multiple groups. P < 0.05 was considered as statistically significant.
Results
C1QTNF1-AS1 is Upregulated in CRC, and Depleted C1QTNF1-AS1 Inhibits the Malignant Processes
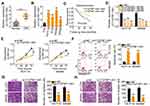
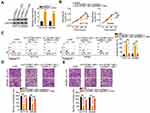
To determine the functions of C1QTNF1-AS1 in CRC, its expression in 71 pairs of CRC tissues and corresponding adjacent normal tissues was determined using RT–qPCR. The data presented a drastic increase in the levels of C1QTNF1-AS1 in CRC tissues when compared to those of corresponding normal tissues (Figure 1A). Furthermore, C1QTNF1-AS1 expression was evidently higher in all four tested CRC cell lines (HCT116, HT29, SW480, and SW620) than in the normal human colon epithelium cell line FHC (Figure 1B). Using the median C1QTNF1-AS1 value in CRC tissues as the cutoff line, all enrolled CRC patients were classified into either C1QTNF1-AS1-low (n = 35) or C1QTNF1-AS1-high (n = 36) expression groups. Increased C1QTNF1-AS1 expression was associated with tumor size (P = 0.026), lymphatic metastasis (P = 0.020) and tumor-node-metastasis (TNM) stage (P = 0.042) (Table 1), but presented none association with gender, age, tumor location, tumor side, N/KRAS gene status, or MSI status (all P > 0.05). In addition, patients with CRC that exhibited high levels of C1QTNF1-AS1 tended to have shorter overall survival than patients with low levels of C1QTNF1-AS1 (Figure 1C, P = 0.033).
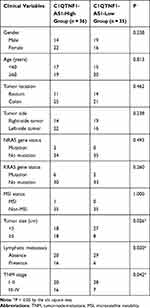 |
Table 1 Correlation Between C1QTNF1-AS1 Expression and Clinical Variables in Patients with CRC |
Since C1QTNF1-AS1 was highly expressed in CRC and closely related to poor prognosis, we speculated that C1QTNF1-AS1 may exert important actions during CRC genesis and progression. HCT116 and SW480 cells were transfected with si-C1QTNF1-AS1 or si-NC. The RT–qPCR data showed that the inhibitory effect of si-C1QTNF1-AS1#3 on C1QTNF1-AS1 expression in both cell lines was more substantial than those of si-C1QTNF1-AS1#1 and si-C1QTNF1-AS1#2 (Figure 1D); hence, si-C1QTNF1-AS1#3 was used in the subsequent experiments. C1QTNF1-AS1 depletion decreased the proliferation of HCT116 and SW480 cells (Figure 1E), as evidenced by the CCK-8 assays. Furthermore, flow cytometric analysis revealed that loss of C1QTNF1-AS1 increased the ratio of apoptotic HCT116 and SW480 cells (Figure 1F). Moreover, interference of C1QTNF1-AS1 resulted in an obvious decrease in the migration (Figure 1G) and invasion (Figure 1H) of HCT116 and SW480 cells. Altogether, these data provide evidence that C1QTNF1-AS1 plays a cancer-promoting role in the oncogenicity of CRC.
C1QTNF1-AS1 Functions as a miR-484 Sponge in CRC Cells
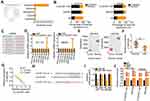
lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/), an lncRNA subcellular localization predictor, identified that C1QTNF1-AS1 was located in the cytoplasm (Figure 2A). Furthermore, subcellular fractionation followed by RT–qPCR confirmed that C1QTNF1-AS1 was primarily distributed in the cytoplasm of HCT116 and SW480 cells (Figure 2B), suggesting that C1QTNF1-AS1 may control gene expression at the posttranscriptional level. It is widely accepted that cytoplasmic lncRNAs can function as miRNA sponges by competitively binding to miRNAs, releasing their downstream target mRNAs.27
Using miRDB, 26 miRNAs had a reverse complementary sequence to C1QTNF1-AS1 (Figure 2C). Among these, eight miRNAs (miR-219b-5p, miR-212-5p, miR-615-3p, miR-488-5p, miR-525-5p, miR-520a-5p, miR-484, and miR-346) were selected as experimental subjects based on their known biological roles in cancer.33–39 To confirm these predictions, RT–qPCR was performed to measure the expression of these miRNAs in HCT116 and SW480 cells after C1QTNF1-AS1 knockdown. MiR-484 expression was increased in HCT116 and SW480 cells upon C1QTNF1-AS1 depletion, whereas the expressions of the other miRNAs was unaffected (Figure 2D). Subsequently, YM500v3 was used to predict the expression status of miR-484 in CRC. MiR-484 was found to be weakly expressed in CRC tissues (Figure 2E). To confirm this prediction, RT–qPCR was performed to determine miR-484 expression in 71 CRC tissue pairs and corresponding adjacent normal tissues. CRC tissues presented lower miR-484 expression than that in adjacent normal tissues (Figure 2F). Notably, Pearson’s correlation coefficient analysis showed that there was a negative correlation between miR-484 and C1QTNF1-AS1 expression in the 71 CRC tissues (Figure 2G; r = −0.7619, P < 0.0001).
Next, the luciferase reporter assay was performed to examine the physical binding relationship between C1QTNF1-AS1 and miR-484 in CRC cells. The wild-type and mutant binding sites between C1QTNF1-AS1 and miR-484 were shown in Figure 2H. The upregulation of miR-484 clearly reduced the luciferase activity of C1QTNF1-AS1-wt harboring the wild-type miR-484 binding sequences; however, the suppression of luciferase activity by miR-484 mimic was abrogated by the mutated binding sequences (Figure 2I). Furthermore, the results of RIP assays showed that C1QTNF1-AS1 and miR-484 were remarkably enriched in the Ago2-containing microribonucleoprotein complexes (Figure 2J). It is generally known that Ago2 is a core component of the RNA-induced silencing complex, and is required for miRNA-mediated gene silencing. MiRNA executes its roles by directly binding to Ago2, and putative miRNA targets can be isolated from this complex after Ago2 co-immunoprecipitation. The results of RIP assay implied the direct interaction between C1QTNF1-AS1 and miR-484 in CRC cells. Altogether, C1QTNF1-AS1 functioned as a miR-484 sponge in CRC cells.
MiR-484 is a Tumor-Inhibiting miRNA and Directly Targets HK2 in CRC Cells
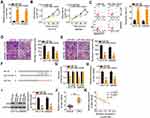
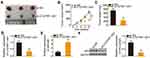
To elucidate the detailed functions of miR-484 in CRC cells, miR-484 mimic or miR-NC was transfected into HCT116 and SW480 cells, and the transfection efficiency was determined using RT–qPCR (Figure 3A). The proliferative abilities of HCT116 and SW480 cells were obviously impaired after transfection with miR-484 mimic (Figure 3B). In addition, the upregulation of miR-484 drastically increased the apoptosis of HCT116 and SW480 cells, as demonstrated by flow cytometry (Figure 3C). Furthermore, cell migration (Figure 3D) and invasion (Figure 3E) strikingly decreased in HCT116 and SW480 cells after miR-484 overexpression.
A potential miR-484 binding site located in the 3′-UTR of HK2 (Figure 3F) was identified using three bioinformatics tools, including miRDB, TargetScan, and StarBase 3.0. A luciferase reporter assay was conducted to further confirm this prediction. MiR-484 mimic or miR-NC together with HK2-wt or HK2-mut reporter plasmids were transfected into HCT116 and SW480 cells. MiR-484 overexpression caused a significant decrease in luciferase activity of HK2-wt in HCT116 and SW480 cells, whereas the HK2-mut-induced luciferase activity did not change in response to miR-484 mimic cotransfection (Figure 3G). Subsequently, the mRNA and protein levels of HK2 in miR-484-overexpressing HCT116 and SW480 cells were determined using RT–qPCR and Western blotting, respectively. As illustrated in Figure 3H and I, HK2 mRNA (Figure 3H) and protein (Figure 3I) levels were lower in HCT116 and SW480 cells transfected with miR-484 mimic than in the control cells. Next, we examined the association between miR-484 and HK2 in CRC tissues. HK2 mRNA expression was elevated in CRC tissues (Figure 3J) and inversely correlated with miR-484 expression (Figure 3K; r = −0.7004, P < 0.0001). These results demonstrate that miR-484 performs tumor-suppressive activities during CRC progression, and HK2 is a direct target of miR-484 in CRC cells.
C1QTNF1-AS1 Plays a Carcinogenic Role by Regulating a miR-484/HK2 Axis
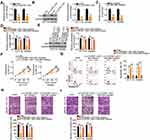
After identifying HK2 as a direct target of miR-484, we wondered whether C1QTNF1-AS1was implicated in the regulation of HK2. Transfection with si-C1QTNF1-AS1 caused a statistically significant decrease (P < 0.01) in HK2 mRNA (Figure 4A) and protein (Figure 4B) levels in HCT116 and SW480 cells. Thereafter, we performed rescue experiments to investigate whether C1QTNF1-AS1 controls HK2 expression in CRC cells by sponging miR-484. First, RT–qPCR analysis was used to verify the efficiency of miR-484 inhibitor transfection (Figure 4C). si-C1QTNF1-AS1, together with miR-484 inhibitor or NC inhibitor, was cotransfected into HCT116 and SW480 cells. The expression levels of HK2 mRNA (Figure 4D) and protein (Figure 4E) were significantly reduced by C1QTNF1-AS1 downregulation, which was recovered by miR-484 inhibition, suggesting that C1QTNF1-AS1positively regulated HK2 expression in CRC cells by sponging miR-484.
To further understand the roles of C1QTNF1-AS1, two rescue experiments were conducted to determine whether C1QTNF1-AS1 exerts an oncogenic role in CRC cells by regulating the miR-484/HK2 axis. First, HCT116 and SW480 cells were transfected with si-C1QTNF1-AS1 in the presence of miR-484 inhibitor or NC inhibitor, and these cells were used in functional rescue experiments. The reduced cell proliferation (Figure 4F) and enhanced cell apoptosis (Figure 4G) by si-C1QTNF1-AS1 were reversed by miR-484 inhibitor cotransfection. Similarly, cell migratory (Figure 4H) and invasive (Figure 4I) abilities were impaired after C1QTNF1-AS1 knockdown but restored after miR-484 inhibition. The HK2 overexpression vector pcDNA3.1-HK2 was also used in rescue experiments, and the overexpression efficiency of pcDNA3.1-HK2 was determined using Western blotting (Figure 5A). Cell proliferation was suppressed by reduced C1QTNF1-AS1 in HCT116 and SW480 cells, whereas this suppressive effect was impaired by the restoration of HK2 (Figure 5B). Strikingly, the rate of HCT116 and SW48 cell apoptosis increased by si-C1QTNF1-AS1 was recovered by pcDNA3.1-HK2 introduction (Figure 5C). Similarly, the loss of C1QTNF1-AS1 hindered HCT116 and SW480 cell migration (Figure 5D) and invasion (Figure 5E), which was rescued by HK2 overexpression. Thus, these results collectively uncovered that C1QTNF1-AS1 played cancer-promoting roles in CRC cells by sponging miR-484 and consequently increasing HK2 expression.
C1QTNF1-AS1 Depletion Inhibits Tumor Growth of CRC Cells in vivo
Lastly, a xenograft tumor model was used to validate the role of C1QTNF1-AS1 in promoting CRC cell growth in vivo. Nude mice were subcutaneously inoculated with SW480 cells stably expressing sh-C1QTNF1-AS1 or sh-NC. A significant decrease in tumor volume (Figure 6A and B) and weight (Figure 6C) was identified in the tumor xenografts that originated from C1QTNF1-AS1-depleted SW480 cells. At 4 weeks post-injection, the nude mice were euthanized and tumor xenografts were collected for molecular detection. Tumors derived from cells with C1QTNF1-AS1 stably knocked down revealed decreased C1QTNF1-AS1expression (Figure 6D) and increased miR-484 (Figure 6E) expression on RT–qPCR analysis. Furthermore, Western blotting indicated that the HK2 protein (Figure 6F) level was downregulated in the tumor xenografts that originated from C1QTNF1-AS1-depleted SW480 cells. Altogether, these results demonstrated the promotive effects of C1QTNF1-AS1 on tumor growth of CRC cells in vivo, which was executed via the regulation of the miR-484/HK2 axis.
Discussion
Over the last few years, lncRNAs have received considerable attention because of their important roles in human cancers.40–42 An increasing number of studies have reported that several lncRNAs are differentially expressed in CRC and contribute to colorectal carcinogenesis and cancer.43–45 LncRNAs promote or suppress diverse malignant processes in CRC, playing oncogenic or tumor-suppressing roles.46–48 A number of lncRNAs have been identified to be dysregulated in CRC, but the regulatory network of lncRNAs continues to remain unclear, requiring further exploration. In this study, we first detected the expression of C1QTNF1-AS1 in CRC and determined the relationship between the clinicopathological parameters and C1QTNF1-AS1 expression among CRC patients. Subsequently, we performed functional analyses to investigate the regulatory roles of C1QTNF1-AS1 during CRC progression. Furthermore, the mechanisms underlying the cancer-promoting actions of C1QTNF1-AS1 in CRC cells were elucidated in detail.
C1QTNF1-AS1 is downregulated in hepatocellular carcinoma.28,29 Functionally, C1QTNF1-AS1 upregulation attenuates cell proliferation, migration, and invasion and induces cell apoptosis in hepatocellular carcinoma.28,29 In addition, ectopic C1QTNF1-AS1 expression hampers the tumor growth of hepatocellular carcinoma in vivo.28,29 However, the expression and roles of C1QTNF1-AS1 have not been fully explored in CRC. Here, RT–qPCR analysis confirmed that C1QTNF1-AS1 expression in CRC tissues increased compared with that in adjacent normal tissues. Interestingly, the analysis of clinical data revealed that an increased level of C1QTNF1-AS1 was related to larger tumor size, higher incidence of lymphatic metastasis, more advanced TNM stage and shorter overall survival, suggesting that C1QTNF1-AS1 may be an oncogenic lncRNA that facilitates CRC progression. A series of experiments uncovered that C1QTNF1-AS1 depletion suppressed CRC cell proliferation, invasion, and migration in vitro. Furthermore, interference of C1QTNF1-AS1 increased cell apoptosis in vitro and reduced cell growth in vivo.
With regard to the mechanism, lncLocator and subcellular fractionation analysis demonstrated that C1QTNF1-AS1 was enriched in the cytoplasm of CRC cells, suggesting that C1QTNF1-AS1 may control gene expression at the posttranscriptional level. Accumulating studies have revealed the extensive interactions among lncRNA-miRNA-mRNA regulatory pathways involving ceRNAs, wherein lncRNAs can recover the expression and roles of miRNAs by sequestering miRNAs.49–51 In our study, we used bioinformatics analysis to screen putative miRNAs that may interact with C1QTNF1-AS1. MiR-484 was identified to potentially interact with C1QTNF1-AS1, and this prediction was confirmed using the luciferase reporter and RIP assays. In addition, RT–qPCR analysis showed that the knockdown of C1QTNF1-AS1 increased miR-484 expression in CRC cells. Furthermore, miR-484 was weakly expressed in CRC and exhibited a reverse correlation with C1QTNF1-AS1 expression in CRC tissues.
Next, the putative targets of miR-484 were explored, and we confirmed that HK2 was a direct target of miR-484 using luciferase reporter assays, RT-PCR analysis, and Western blotting. The regulatory relationship between C1QTNF1-AS1, miR-484, and HK2 was then evaluated in detail. Our results showed that C1QTNF1-AS1 downregulation reduced HK2 expression in CRC cells at both mRNA and protein levels. In subsequent rescue assays, we found that miR-484 inhibition could partially abrogate the regulatory actions of C1QTNF1-AS1 knockdown on HK2 expression in CRC cells. Overall, these findings revealed that C1QTNF1-AS1 is a type of ceRNA that functions as an miR-484 sponge and thereby increases HK2 expression.
MiR-484 has been reported to be aberrantly expressed in a variety of human cancers, including CRC.52 In CRC, miR-484 was downregulated and closely correlated with the tumor stage.53 HK2, a key metabolic enzyme,54 was identified as the direct downstream target of miR-484 in CRC cells. In this study, miR-484 inhibition or HK2 restoration diminished the impacts of C1QTNF1-AS1 deficiency on the aggressiveness of CRC cells, suggesting that C1QTNF1-AS1partially executed its cancer-promoting actions in CRC cells by modulating a miR-484/HK2 axis. Therefore, these results identified a novel ceRNA pathway in CRC involving C1QTNF1-AS1, miR-484, and HK2. The C1QTNF1-AS1/miR-484/HK2 pathway may contribute to CRC pathogenesis.
In the study, a total of 6 mice was used in xenograft tumor model, and each group contained three nude mice. The number of mice per group was low, and it was a limitation of our study. We will resolve it in the near future.
Conclusion
In conclusion, this study revealed the abnormally high expression of C1QTNF1-AS1 in CRC and its clinical relevance. C1QTNF1-AS1 drove the progression of CRC by adsorbing miR-484 and consequently upregulating HK2. The C1QTNF1-AS1/miR-484/HK2 pathway may be a potential therapeutic target for the diagnosis and treatment of CRC.
Ethics Approval and Informed Consent
The present study was approved by the Ethics Committee of Jilin Cancer Hospital and conducted in accordance with the Declaration of Helsinki. Moreover, written informed consent forms were collected from all participants. The animal experimental protocol was approved by the Ethics Committee of Jilin Cancer Hospital and complied with the Animal Protection Law of the People’s Republic of China-2009 for experimental animals.
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289.
3. Guraya SY. Pattern, stage, and time of recurrent colorectal cancer after curative surgery. Clin Colorectal Cancer. 2019;18(2):e223–e228. doi:10.1016/j.clcc.2019.01.003
4. Loree JM, Kopetz S. Recent developments in the treatment of metastatic colorectal cancer. Ther Adv Med Oncol. 2017;9(8):551–564. doi:10.1177/1758834017714997
5. Banerjee A, Pathak S, Subramanium VD, Murugesan R, Verma RS. Strategies for targeted drug delivery in treatment of colon cancer: current trends and future perspectives. Drug Discov Today. 2017;22(8):1224–1232. doi:10.1016/j.drudis.2017.05.006
6. Rees M, Tekkis PP, Welsh FK, O’Rourke T, John TG. Evaluation of long-term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann Surg. 2008;247(1):125–135.
7. Donadon M, Ribero D, Morris-Stiff G, Abdalla EK, Vauthey JN. New paradigm in the management of liver-only metastases from colorectal cancer. Gastrointest Cancer Res. 2007;1(1):20–27.
8. Soleimani A, Pashirzad M, Avan A, Ferns GA, Khazaei M, Hassanian SM. Role of the transforming growth factor-beta signaling pathway in the pathogenesis of colorectal cancer. J Cell Biochem. 2019;120(6):8899–8907.
9. Soleimani A, Rahmani F, Ferns GA, Ryzhikov M, Avan A, Hassanian SM. Role of the NF-kappaB signaling pathway in the pathogenesis of colorectal cancer. Gene. 2020;726:144132. doi:10.1016/j.gene.2019.144132
10. Cheng KJ, Alshawsh MA, Mejia Mohamed EH, Thavagnanam S, Sinniah A, Ibrahim ZA. HMGB1: an overview of its versatile roles in the pathogenesis of colorectal cancer. Cell Oncol. 2020;43(2):177–193.
11. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606
12. Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24. doi:10.1016/j.addr.2015.05.012
13. Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148–64167. doi:10.18632/oncotarget.11637
14. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi:10.1016/j.bbagrm.2014.08.012
15. Zhang Y, Li Z, Lan Z. Silencing UNC5B antisense lncRNA 1 represses growth and metastasis of human colon cancer cells via raising miR-622. Artif Cells Nanomed Biotechnol. 2020;48(1):60–67. doi:10.1080/21691401.2019.1699809
16. Ge H, Yan Y, Yue C, Liang C, Wu J. Long noncoding RNA LINC00265 targets EGFR and promotes deterioration of colorectal cancer: a comprehensive study based on data mining and in vitro validation. Onco Targets Ther. 2019;12:10681–10692.
17. Sun W, Ren S, Li R, Zhang Q, Song H. LncRNA, a novel target biomolecule, is involved in the progression of colorectal cancer. Am J Cancer Res. 2019;9(11):2515–2530.
18. Chen S, Liu Y, Wang Y, Xue Z. LncRNA CCAT1 promotes colorectal cancer tumorigenesis via A miR-181b-5p/TUSC3 axis. Onco Targets Ther. 2019;12:9215–9225. doi:10.2147/OTT.S216718
19. Yang B, Zhou SN, Tan JN, et al. Long non-coding RNA STARD13-AS suppresses cell proliferation and metastasis in colorectal cancer. Onco Targets Ther. 2019;12:9309–9318. doi:10.2147/OTT.S217094
20. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
21. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi:10.1038/nrg1379
22. Yang H, Ren J, Bai Y, Jiang J, Xiao S. MicroRNA-518-3p suppresses cell proliferation, invasiveness, and migration in colorectal cancer via targeting TRIP4. Biochem Cell Biol. 2020;98(5):575–582. doi:10.1139/bcb-2019-0442
23. Zhou F, Tang D, Xu Y, et al. Identification of microRNAs and their endonucleolytic cleavaged target mRNAs in colorectal cancer. BMC Cancer. 2020;20(1):242. doi:10.1186/s12885-020-06717-4
24. Hong YG, Xin C, Zheng H, et al. miR-365a-3p regulates ADAM10-JAK-STAT signaling to suppress the growth and metastasis of colorectal cancer cells. J Cancer. 2020;11(12):3634–3644. doi:10.7150/jca.42731
25. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
26. Ye Y, Shen A, Liu A. Long non-coding RNA H19 and cancer: a competing endogenous RNA. Bull Cancer. 2019;106(12):1152–1159. doi:10.1016/j.bulcan.2019.08.011
27. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20(22).
28. Li H, Zhang B, Ding M, et al. C1QTNF1-AS1 regulates the occurrence and development of hepatocellular carcinoma by regulating miR-221-3p/SOCS3. Hepatol Int. 2019;13(3):277–292. doi:10.1007/s12072-019-09944-5
29. Han W, Yu G, Meng X, et al. Potential of C1QTNF1-AS1 regulation in human hepatocellular carcinoma. Mol Cell Biochem. 2019;460(1–2):37–51. doi:10.1007/s11010-019-03569-w
30. Xu T, Lei T, Li SQ, Mai EH, Ding FH, Niu B. DNAH17-AS1 promotes pancreatic carcinoma by increasing PPME1 expression via inhibition of miR-432-5p. World J Gastroenterol. 2020;26(15):1745–1757. doi:10.3748/wjg.v26.i15.1745
31. Chen Z, Wu H, Zhang Z, Li G, Liu B. LINC00511 accelerated the process of gastric cancer by targeting miR-625-5p/NFIX axis. Cancer Cell Int. 2019;19:351. doi:10.1186/s12935-019-1070-0
32. Cheng R, Lu X, Xu C, Zhang F, Zhang G. SNHG11 contributes to NSCLC cell growth and migration by targeting miR-485-5p/BSG axis. Biomed Pharmacother. 2020;128:110324. doi:10.1016/j.biopha.2020.110324
33. Lv ZD, Yang DX, Liu XP, et al. MiR-212-5p suppresses the epithelial-mesenchymal transition in triple-negative breast cancer by targeting Prrx2. Cel Physiol Biochem. 2017;44(5):1785–1795. doi:10.1159/000485785
34. Sun Y, Wang J, Pan S, et al. LINC00657 played oncogenic roles in esophageal squamous cell carcinoma by targeting miR-615-3p and JunB. Biomed Pharmacother. 2018;108:316–324. doi:10.1016/j.biopha.2018.09.003
35. Arnold J, Engelmann JC, Schneider N, Bosserhoff AK, Kuphal S. miR-488-5p and its role in melanoma. Exp Mol Pathol. 2020;112:104348. doi:10.1016/j.yexmp.2019.104348
36. Chen M, Liu LX. MiR-525-5p repressed metastasis and anoikis resistance in cervical cancer via blocking UBE2C/ZEB1/2 signal axis. Dig Dis Sci. 2019.
37. Liu X, Song J, Kang Y, Wang Y, Chen A. Long noncoding RNA SOX21-AS1 regulates the progression of triple-negative breast cancer through regulation of miR-520a-5p/ORMDL3 axis. J Cell Biochem. 2020.
38. Shen Y, Qi L, Li Y, et al. The downregulation of lncRNA PGM5-AS1 inhibits the proliferation and metastasis via increasing miR-484 expression in colorectal cancer. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2019.3059
39. Li Y, Xu J, Zhang J, Zhang J, Zhang J, Lu X. MicroRNA-346 inhibits the growth of glioma by directly targeting NFIB. Cancer Cell Int. 2019;19:294. doi:10.1186/s12935-019-1017-5
40. Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19(1):77. doi:10.1186/s12943-020-01188-4
41. Connerty P, Lock RB, de Bock CE. Long non-coding RNAs: major regulators of cell stress in cancer. Front Oncol. 2020;10:285. doi:10.3389/fonc.2020.00285
42. Agostini M, Ganini C, Candi E, Melino G. The role of noncoding RNAs in epithelial cancer. Cell Death Discov. 2020;6:13.
43. Galamb O, Bartak BK, Kalmar A, et al. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J Gastroenterol. 2019;25(34):5026–5048. doi:10.3748/wjg.v25.i34.5026
44. Zhao K, Ye Z, Li Y, et al. LncRNA FTX contributes to the progression of colorectal cancer through regulating miR-192-5p/EIF5A2 axis. Onco Targets Ther. 2020;13:2677–2688. doi:10.2147/OTT.S241011
45. Liu J, Qian J, Mo Q, Tang L, Xu Q. LncRNA NR2F2-AS1 silencing induces cell cycle arrest in G0/G1 Phase via downregulating cyclin D1 in colorectal cancer. Cancer Manag Res. 2020;12:1835–1843. doi:10.2147/CMAR.S221996
46. Gao Q, Zhou R, Meng Y, et al. Long noncoding RNA CMPK2 promotes colorectal cancer progression by activating the FUBP3-c-Myc axis. Oncogene. 2020;39(19):3926–3938. doi:10.1038/s41388-020-1266-8
47. Hong S, Yan Z, Song Y, Bi M, Li S. LncRNA AGAP2-AS1 augments cell viability and mobility, and confers gemcitabine resistance by inhibiting miR-497 in colorectal cancer. Aging. 2020;12(6):5183–5194. doi:10.18632/aging.102940
48. Jiang HY, Wang ZJ. ADPGK-AS1 promotes the progression of colorectal cancer via sponging miR-525 to upregulate FUT1. Eur Rev Med Pharmacol Sci. 2020;24(5):2380–2386.
49. Zheng X, Dong S, Sun L, Xu J, Liu J, Hao R. LncRNA LINC00152 promotes laryngeal cancer progression by sponging MiR-613. Open Med. 2020;15:240–248.
50. Ma YL, Wang CY, Guan YJ, Gao FM. Long noncoding RNA ROR promotes proliferation and invasion of colorectal cancer by inhibiting tumor suppressor gene NF2 through interacting with miR-223-3p. Eur Rev Med Pharmacol Sci. 2020;24(5):2401–2411.
51. Liu X, Xie S, Zhang J, Kang Y. Long noncoding RNA XIST contributes to cervical cancer development through targeting miR-889-3p/SIX1 axis. Cancer Biother Radiopharm. 2020.
52. Xie S, Ge Q, Wang X, Sun X, Kang Y. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle. 2018;17(2):154–161. doi:10.1080/15384101.2017.1407895
53. Lu X, Lu J. The significance of detection of serum miR-423-5p and miR-484 for diagnosis of colorectal cancer. Clin Lab. 2015;61(1–2):187–190. doi:10.7754/Clin.Lab.2014.140625
54. Zhou P, Chen WG, Li XW. MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. Am J Cancer Res. 2015;5(6):2056–2063.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.