Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA BLACAT1 Promotes the Tumorigenesis of Gastric Cancer by Sponging microRNA-149-5p and Targeting KIF2A
Authors Wang Z, Liu X, Liu X, Niu D
Received 21 April 2020
Accepted for publication 9 July 2020
Published 30 July 2020 Volume 2020:12 Pages 6629—6640
DOI https://doi.org/10.2147/CMAR.S258178
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
This paper has been retracted.
Zhengkun Wang,1 Xichun Liu,1 Xiaolei Liu,2 Dongguang Niu1
1Department of Gastrointestinal Surgery, Affiliated Hospital of Qingdao University, Qingdao City, Shandong Province 266000, People’s Republic of China; 2Department of General Surgery, Affiliated Hospital of Qingdao University, Qingdao City, Shandong Province 266000, People’s Republic of China
Correspondence: Dongguang Niu Tel +86-13963999533
Email [email protected]
Objective: Gastric cancer (GC) is a gastrointestinal tumor. This study is aimed to explore the regulatory mechanism of long non-coding RNA BLACAT1 (BLACAT1)/microRNA-149-5p (miR-149-5p)/KIF2A cascade on GC.
Methods: The expression of BLACAT1, miR-149-5p and KIF2A in GC was detected by qRT-PCR. The proliferation, migration and invasion of GC cells in vitro were analyzed by MTT, wound-healing and transwell assay, respectively. The xenograft tumor model was constructed in nude mice to confirm the inhibition effect of BLACAT1 knockdown on GC in vivo. Then, dual-luciferase reporter assay was used to detect the interactions among BLACAT1, miR-149-5p and KIF2A. Western blot assay was performed to determine the protein expression of KIF2A.
Results: The expression of BLACAT1 and KIF2A was up-regulated in GC, but miR-149-5p expression was down-regulated. Silencing of BLACAT1 retarded the proliferation, migration and invasion of GC cells in vitro and the growth of tumor xenograft in vivo. Moreover, BLACAT1 acted as the molecular sponge of miR-149-5p to up-regulate KIF2A expression. At last, feedback experiments suggested that BLACAT1 accelerated the proliferation, migration and invasion of GC cells by regulating miR-149-5p/KIF2A axis.
Conclusion: BLACAT1 facilitated the tumorigenesis of GC through regulating miR-149-5p/KIF2A axis, which indicated BLACAT1/miR-149-5p/KIF2A cascade may be a new therapeutic target.
Keywords: gastric cancer, long non-coding RNA BLACAT1, microRNA-149-5p, KIF2A, proliferation
Introduction
Gastric cancer (GC) is a gastrointestinal tumor and the third cancer-related death cause worldwide.1,2 GC is asymptomatic in early stages, and diagnosed at an advanced stage in most patients.3 The mortality of GC is high and successful therapeutic strategies are limited.4 Thus, it is indispensable to explore critical molecules involved in GC cell tumorigenesis and develop new effective therapies.
Long noncoding RNAs (lncRNAs) take part in biological processes of gastrointestinal malignancies, such as colorectal cancer,5 esophageal cancer6 and GC.7 LncRNA bladder cancer-associated transcript 1 (BLACAT1) participates in multiple biological processes of cancers. For instance, BLACAT1 exhibits tumor pro-oncogenic activity and is a negative prognostic factor in colorectal cancer.8 BLACAT1 contributes to tumorigenesis of cervical cancer cells.9 BLACAT1 high-expression acts as a poor outcome biomarker for overall survival in small-cell lung cancer.10 Importantly, BLACAT1 elevates the oxaliplatin-resistance of GC by enhancing ABCB1 protein expression through inhibiting miR-361.11 However, the detailed mechanism of BLACAT1 on the occurrence and development of GC needs to be further explored. Some microRNAs (miRNAs) have anti-cancer effect on GC. For example, miR-449 functions as anti-onco-miR in human GC.12 miR-126 retards cell growth and is down-regulated in GC.13 Researchers have indicated that miR-149 is associated with the processes of GC. Wang et al have proved that miR-149 attenuates cell-cycle progression and proliferation in GC.14 Zhang et al have showed that miR-149-5p reduces the proliferation rate of GC cells.15 Notably, lncRNA GACAT1 increases GC cell growth, invasion and migration via inhibiting miR-149.16 However, it is still unknown whether the suppressive effect of miR-149 on the development of GC is regulated by BLACAT1.
Kinesin superfamily proteins 2A (KIF2A) belongs to KIFs, and serves as a role of microtubule stabilizer.17 KIF2A takes part in the progression of several malignancies, including ovarian cancer,18 glioma19 and colorectal cancer.20 Additionally, up-regulation of KIF2A is associated with poor prognoses and clinicopathological features in GC patients.21 Nevertheless, the potential regulatory mechanism of BLACAT1 related to KIF2A in GC remains undefined.
Herein, we determined the expression and functional role of BLACAT1 in GC. The regulatory relationship between BLACAT1 and miR-149-5p was further evaluated. Then, we explored whether KIF2A was a target of miR-149-5p. A BLACAT1/miR-149-5p/KIF2A cascade in GC pathogenesis was identified. Our research may uncover a novel therapeutic target for GC.
Materials and Methods
Tissue Samples
In total 52 GC patients (26 males and 26 females) were collected from our hospital from January 2017 to January 2019. Patients had not received preoperative systemic or local treatment before this study. Tumor tissues and adjacent tissues were collected. We separated our patient cohort according to the median BLACAT1 expression into high BLACAT1 (n = 26) and low BLACAT1 groups (n = 26). This study was permitted by our hospital ethics committee, and the informed consents were obtained from each patient.
Cell Culture
Four human GC cell lines (AGS, MNK-45, MGC803 and SNU-5) and human normal gastric epithelial cell line GES-1 were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, Invitrogen) at 37°C.
Cell Transfection
The siRNA-negative control (si-NC), si-BLACAT1-1, si-BLACAT1-2, miR-NC, miR-149-5p mimics, miR-149-5p inhibitor, pcDNA3.1-NC (pcDNA-NC) and pcDNA3.1-KIF2A (pcDNA-KIF2A) were synthesized by Shanghai Genepharma (Shanghai, China). AGS and MKN45 cells grown to 80% confluence were transfected or co-transfected with the above agents using Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s protocol.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using the TRIzol reagent (Invitrogen) and was reverse-transcribed into cDNA by PrimeScript RT reagent kit (Takara, Otsu, Japan). PCR reaction was performed on ABI 7500HT Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) with the following conditions: 95°C for 5 min, 40 cycles of 72°C for 34 s and 60°C for 20 s. Relative expression was calculated by the 2−ΔΔCt method. GAPDH, U6 or β-actin were used for the normalization of BLACAT1, miR-149-5p and KIF2A, respectively. The primer sequences are shown in Table 1.
 |
Table 1 Primer Sequences |
Western Blot
AGS and MKN45 cells were lysed by ice-cold lysis buffer to obtain total proteins. Protein samples were size-fractionated in SDS-PAGE, and transferred to polyvinylidene fluoride membranes. Then, membranes were blocked with 5% skimmed milk and incubated overnight at 4°C with primary antibody anti-KIF2A (1:1000, SAB2101262MSDS, Sigma, St. Louis, MO, USA). Afterwards, the membranes were probed with HRP conjugate secondary antibody (1:5000, 12–348MSDS, Sigma) for 1 h at 25°C. The protein blots were visualized by ECL chemiluminescent substrate reagent kit (Invitrogen), and quantified by ImageLab software (Bio-Rad, Hercules, CA, USA). β-actin (1:5000, SAB2701711MSDS, Sigma) was introduced as the internal reference.
MTT Assay
AGS and MKN45 cells were seeded into 96-well plates (2 × 103 cells/well) and cultured with 5% CO2 at 37°C. Cell proliferation was measured using the MTT cell proliferation assay kit (Sigma) according to the guidelines.
Wound-Healing Assay
AGS and MKN45 cells were seeded into a 6-well plate and cultured until 80% confluence. A sterile pipette tip was used to draw a vertical line on the cell surface. Cells were then cultured in DMEM without FBS for 24 h. The scratch width was measured at 0 and 24 h post-incubation, respectively. Cells were observed under a light microscope and the wound-healing rate was calculated as: (1–scratch width at 24 h/scratch width at 0 h) × 100%.
Transwell Assay
The transwell assay was used to measure the cell invasion by using transwell chambers (8 nm pore size) (Corning Inc., Corning, NY, USA). AGS and MKN45 cells (2 × 105) in serum-free medium were placed to upper chambers pre-coated with matrigel (Sigma). Medium with 10% FBS was added to the lower chambers. Cells remaining in the upper chamber were wiped with cotton swab after 48 h of incubation at 37°C, while cells in lower chambers were fixed in methanol, and stained with 0.1% crystal violet. Five random fields were selected in each well for counting under an inverted microscope (Olympus, Tokyo, Japan).
Tumor Xenograft in Nude Mice
Female BALB/c nude mice (weighing 20 ± 2g) were purchased from Cavens Lab, Ltd (Changzhou, China) and fed in sterile environment at 25°C. Si-BLACAT1 or si-NC was integrated into lentiviral vector and then transfected into AGS cells. After that, the transfected AGS cells (2 × 106 cells/100 ul, s.c.) were injected into the right flanks of the nude mice (n = 5). Tumor volumes were measured every other week. Four weeks later, mice were anesthetized with pentobarbital sodium (50 mg/kg) and then sacrificed by cervical dislocation. The tumor xenograft was separated from mice and weighted. The study was performed in the Animal Experimental Center of our hospital and obtained the approval of ethical committee in our hospital.
Dual-Luciferase Reporter Assay
The potential binding sites of miR-149-5p and BLACAT1 or miR-149-5p and KIF2A were predicted by Starbase or TargetScan, respectively. BLACAT1 with WT or MUT miR-149-5p-binding sites were generated and fused to the psiCHECK-2 vector (Promega, Madison, WI, USA). The 3ʹ-UTR of KIF2A containing the predicted miR-149-5p targeting site (WT), and MUT KIF2A were amplified and cloned into the psiCHECK-2 vector. Then, BLACAT1-Wt and BLACAT1-Mut vectors were co-transfected with miR-NC or miR-149-5p mimics into AGS and MKN45 cells. KIF2A-Wt and KIF2A-Mut vectors were co-transfected with miR-NC + pcDNA-NC, miR-149-5p mimics + pcDNA-NC or miR-149-5p mimics + pcDNA-BLACAT1 into AGS and MKN45 cells. Cell transfection was performed using Lipofectamine 3000 following the guidelines.
Statistical Analysis
Statistical analysis was performed on GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). Data were displayed as mean ± standard deviation (SD). The differences between two groups or among multiple groups were assessed by Student’s t-test or one-way ANOVA followed by Tukey’s post hoc test. The correlation significance was determined by Pearson correlation analysis. A P value <0.05 was considered statistically significant.
Results
The Expression of BLACAT1 Was Increased in GC Tissues and Cells
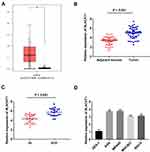
Through TCGA database, BLACAT1 expression in stomach adenocarcinoma (STAD) and normal tissues was analyzed. The results showed that BLACAT1 expression was increased in STAD tissues in contrast to that in normal tissues (P < 0.05) (Figure 1A). Then we measured BLACAT1 expression in tissues of GC patients by qRT-PCR. The BLACAT1 expression in tumor tissues was obviously increased by contrast to the adjacent tissues in GC patients (P < 0.001) (Figure 1B). And its level in tissues in WHO grade III/IV was visibly higher than that in WHO grade I/II (P < 0.001) (Figure 1C). The BLACAT1 expression was visibly enhanced in AGS, MKN45, MGC803 and SNU-5 cells by contrast to the GES-1 cells (P < 0.01) (Figure 1D). AGS and MKN45 cells were used for the subsequent assays. As shown in Table 2, BLACAT1 expression had no connections with age, gender, diameter and resection degree in GC patients, while the BLACAT1 expression was positively correlated with WHO grade (P < 0.01) and TNM stage (P < 0.05).
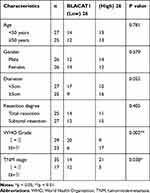 |
Table 2 Correlation Between BLACAT1 Expression and Clinicopathological Features in Gastric Cancer (GC) |
Silencing of BLACAT1 Inhibited the Proliferation, Migration and Invasion of GC Cells in vitro and the Growth of Tumor Xenograft in vivo
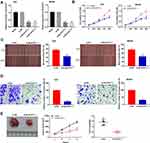
To evaluate the effect of BLACAT1 on tumorigenesis of GC, BLACAT1 was silenced by the transfection of si-BLACAT1-1 and −2 (P < 0.01) (Figure 2A). Then, si-BLACAT1-1 was used for the subsequent assays. MTT assay discovered that si-BLACAT1-1 significantly decreased the proliferation of AGS and MKN45 cells at 96 h post-culturing (P < 0.01) (Figure 2B). Wound-healing assay and transwell assay disclosed that the wound-healing rate and number of invasion GC cells were markedly reduced in the si-BLACAT1-1 group compared with the si-NC group (P < 0.01) (Figure 2C and D). Additionally, we also investigated the effect of BLACAT1 knockdown on the growth of tumor xenograft in vivo. Four weeks later, the results demonstrated that in comparison to the si-NC group, tumor volume (P < 0.05) and tumor weight (P < 0.01) were both distinctly decreased in the si- BLACAT-1 group (Figure 2E).
BLACAT1 Was Negatively Correlated with miR-149-5p
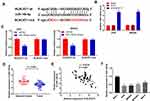
Starbase was used to predict a binding site of miR-149-5p on the 3ʹ UTR of BLACAT1 (Figure 3A). Silencing of BLACAT1 markedly enhanced the miR-149-5p expression in AGS and MKN45 cells (P < 0.01) (Figure 3B). Next, the relative luciferase activity was obviously declined in AGS and MKN45 cells co-transfected with miR-149-5p mimics and BLACAT1 Wt by contrast to the cells co-transfected with miR-NC and BLACAT1 Wt (P < 0.01, Figure 3C). Moreover, miR-149-5p expression in tumor tissues was notably decreased by contrast to the adjacent normal tissues (P < 0.001) (Figure 3D). There was a negative correlation between BLACAT1 and miR-149-5p expression in GC tissues (N = 52, r = −0.5319, P < 0.001) (Figure 3E). Furthermore, miR-149-5p expression was obviously lower in AGS, MKN45, MGC803 and SNU-5 cells than that in GES-1 cells (P < 0.01) (Figure 3F).
miR-149-5p Inhibited Proliferation, Migration and Invasion of GC Cells
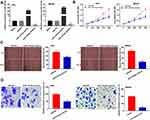
To determine the biological function of miR-149-5p in GC, miR-149-5p was overexpressed by the transfection of miR-149-5p mimics, and inhibited by the transfection of miR-149-5p inhibitor (P < 0.01) (Figure 4A). miR-149-5p up-regulation markedly attenuated the proliferation of AGS and MKN45 cells at 96 h post-culturing (P < 0.01) (Figure 4B). Additionally, the wound-healing rate and number of invasion GC cells were lower in the miR-149-5p mimics group than those in the miR-NC group (P < 0.01) (Figure 4C and D).
BLACAT1 Positively Regulated KIF2A Expression via Binding miR-149-5p
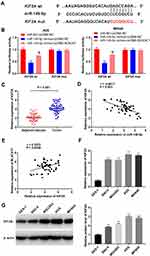
TargetScan predicted a binding site for miR-149-5p on the 3ʹ UTR of KIF2A (Figure 5A). The relative luciferase activity was visibly decreased in AGS and MKN45 cells co-transfected with miR-149-5p mimics + pcDNA-NC and KIF2A Wt compared with the cells co-transfected with miR-NC + pcDNA-NC and KIF2A Wt (P < 0.01). Overexpression of BLACAT1 reversed the reducing effect of miR-149-5p mimics on the relative luciferase activity (P < 0.05) (Figure 5B). Furthermore, the KIF2A expression in tumor tissues was markedly increased by contrast to the adjacent tissues in GC patients (P < 0.001) (Figure 5C). The expression of KIF2A and miR-149-5p was negatively correlated in GC tissues (N = 52, r = −0.6011, P < 0.001) (Figure 5D), but the expression of KIF2A and BLACAT1 was positively correlated (N = 52, r = 0.3872, P = 0.0046) (Figure 5E). Additionally, the KIF2A expression was obviously higher in AGS, MKN45, MGC803 and SNU-5 cells than that in GES-1 cells (P < 0.01) (Figure 5F). Western blot assay demonstrated that the protein level of KIF2A in GC cells (SNU-5, MGC803, AGS and MKN45) was distinctly elevated in comparison to that in GES-1 cells (P < 0.01) (Figure 5G).
Knockdown of BLACAT1 Attenuated the Proliferation, Migration and Invasion of GC Cells via Targeting miR-149-5p/KIF2A Axis
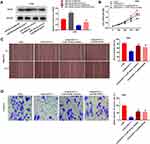
Western blot assay revealed that in AGS cells, miR-149-5p overexpression down-regulated the protein expression of KIF2A and BLACAT1 overexpression up-regulated the protein expression of KIF2A (P < 0.01). At the same time, BLACAT1 reversed the suppressive effect of miR-149-5p on KIF2A protein expression (P < 0.05) (Figure 6A). The proliferation, wound-healing rate and invasion rate of AGS cells were obviously declined in the si-BLACAT1-1 group by contrast to the si-NC group (P < 0.01). Overexpression of KIF2A or inhibition of miR-149-5p strikingly reversed the reduction effect of si-BLACAT1-1 on the proliferation, migration and invasion of AGS cells (P < 0.05) (Figure 6B-D).
Discussion
Dysregulation of lncRNAs involves in the tumorigenesis of GC.22,23 LncRNAs such as lncRNA HOX11-AS,24 lncRNA PVT125 and lncRNA Sox2ot26 are all up-regulated in patients with GC. Here, BLACAT1 expression was enhanced in GC tissues and cells, and was correlated with TNM stage in GC patients. The function of BLACAT1 was similar to some previously reported lncRNAs. Yao et al have pointed out that lncRNA CASC15 expression is related to the TNM stage in GC patients.27 Peng et al have proved that high-expressed lncRNA EGOT is associated with metastasis and TNM stage in GC patients.28 In addition, we also demonstrated that BLACAT1 expression was correlated with WHO grade. Therefore, our data implied that BLACAT1 might act as a pathogenic-factor role in GC.
Prior researches have displayed that BLACAT1 takes part in the biological processes of various cancers.29 For instance, BLACAT1 accelerates the cell proliferation and invasion in glioma.30 BLACAT1 contributes to cell growth and inhibits cell cycle arrest in ovarian cancer.31 In the present study, we found that BLACAT1 knockdown suppressed the proliferation, migration and invasion of GC cells in vitro. Consistent with our results, a previous study has revealed that BLACAT1 knockdown attenuates the proliferation, migration and invasion of GC cells in vitro.30 However, the prior study only investigated the inhibitory effect of BLACAT1 knockdown on GC cells in vitro. Our results further indicated that BLACAT1 knockdown also inhibited the growth of tumor xenograft in vivo. The above results suggested that BLACAT1 knockdown retarded the progression of GC.
Numerous studies have proved that miR-149-5p serves as anti-onco-miR in multiple cancers. Jin et al have shown that the expression of miR-149-5p is downregulated in renal cell carcinoma and overexpression of miR-149-5p has an anti-tumor effect on the progression of renal cell carcinoma.32 Chen et al have reported that miR-149-5p expression is distinctly reduced in melanoma and miR-149-5p up-regulation can hinder the cell proliferation of melanoma.33 Notably, miR-149 expression is markedly decreased in GC tissues and overexpression of miR-149 diminishes the viability of GC cells.34 In the current study, miR-149-5p expression was declined in GC tissues and cells, and miR-149-5p overexpression attenuated the proliferation, migration and invasion of GC cells. The results implied that miR-149-5p might serve as a suppressor in GC. LncRNAs can function as ceRNAs or molecular sponges in modulating miRNAs in the progression of GC.35,36 BLACAT1 has been reported to act as an onco-lncRNA by targeting miRNAs. For example, BLACAT1 interacts with miR-485-5p to accelerate the tumorigenesis of hepatocellular carcinoma.37,38 BLACAT1 contributes to the glioma development through binding to miR-605-3p.39 BLACAT1 enhances the cell survival and metastasis of breast cancer by miR-150-5p/CCR2 axis.40 We suspected that BLACAT1 mediated the progression of GC by similar mechanisms above. In this study, miR-149-5p was proved to be the target of BLACAT1. At the same time, down-regulation of miR-149-5p reversed the inhibitory effect of BLACAT1 knockdown on GC cells. The above results further indicated that BLACAT1 might promote tumorigenesis of GC by modulating miR-149-5p.
KIF2A has been reported to be taken part in the occurrence and development of GC.41–43 KIF2A acts as a molecular target for the therapeutics of squamous cell carcinoma of the oral tongue.41 Knockdown of KIF2A reduces the cell viability of diffuse large B cell lymphoma.42 Importantly, KIF2A is increased in GC and KIF2A silencing can retard the proliferation and invasion of GC cells.43 In this study, KIF2A expression was increased in GC tumor, which is consistent with the previous studies.43–45 The results suggested that KIF2A might be a pathogenic factor in GC. KIF2A usually exerts its role in regulating the tumorigenesis as a target of miRNAs, such as miR-206a-KIF2A in ovarian cancer and miR-451a-KIF2A in lung squamous cell carcinoma.18,46 In the current study, KIF2A was a target of miR-149-5p and was negatively regulated by miR-149-5p. We suspected that miR-149-5p was involved in the development of GC by targeting KIF2A. Additionally, the KIF2A expression was positively related to BLACAT1 in GC. We further assumed that BLACAT1 might serve as miR-149-5p sponge to inhibit KIF2A expression and then elevate KIF2A expression in GC. To further validate these conjectures, the feedback verification experiment showed that KIF2A up-regulation and miR-149-5p down-regulation both reversed the anti-tumor effect of BLACAT1 silencing on GC cells. To sum up, we indicated that BLACAT1 interacted with miR-149-5p to promote the tumorigenesis of GC via targeting KIF2A.
Conclusions
In conclusion, BLACAT1 expression was up-regulated in GC tissues and cells. miR-149-5p was targeted by BLACAT1 and KIF2A was targeted by miR-149-5p. BLACAT1 up-regulated KIF2A expression by interacting with miR-149-5p, and then promoted the proliferation, migration and invasion of GC cells. Thus, BLACAT1 may be a promising therapeutic target for GC.
Abbreviations
GC, gastric cancer; BLACAT1, long non-coding RNA BLACAT1; miR-149-5p, microRNA-149-5p; lncRNAs, long noncoding RNAs; miRNAs, microRNAs; si-NC, siRNA-negative control; pcDNA-NC, pcDNA3.1-NC; qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation.
Data Sharing Statement
The data in this study can be obtained from the corresponding author upon reasonable request.
Ethics and Consent
This study was permitted by Affiliated Hospital of Qingdao University ethics committee, and informed consents were obtained from each patient. All participants agreed to the study being published.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
All authors declare that there are no competing interests.
References
1. Venerito M, Vasapolli R, Rokkas T, Delchier JC, Malfertheiner P. Helicobacter pylori, gastric cancer and other gastrointestinal malignancies. Helicobacter. 2017;22(Suppl):1. doi:10.1111/hel.12413
2. Figueiredo C, Costa S, Karameris A, Machado JC. Pathogenesis of gastric cancer. Helicobacter. 2015;20(Suppl 1):30–35. doi:10.1111/hel.12254
3. Song H, Zhu J, Lu D. Molecular-targeted first-line therapy for advanced gastric cancer. Cochrane Database Syst Rev. 2016;7:CD011461.
4. Duraes C, Almeida GM, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Arch. 2014;464(3):367–378. doi:10.1007/s00428-013-1533-y
5. Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181.
6. Jiao C, Song Z, Chen J, et al. lncRNA-UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer. Oncol Rep. 2016;36(5):2960–2966. doi:10.3892/or.2016.5121
7. Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi:10.18632/oncotarget.1913
8. Su J, Zhang E, Han L, et al. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis. 2017;8(3):e2665. doi:10.1038/cddis.2017.83
9. Shan D, Shang Y, Hu T. Long noncoding RNA BLACAT1 promotes cell proliferation and invasion in human cervical cancer. Oncol Lett. 2018;15(3):3490–3495. doi:10.3892/ol.2018.7773
10. Chen W, Hang Y, Xu W, et al. BLACAT1 predicts poor prognosis and serves as oncogenic lncRNA in small-cell lung cancer. J Cell Biochem. 2018.
11. Wu X, Zheng Y, Han B, Dong X. Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed Pharmacother. 2018;99:832–838. doi:10.1016/j.biopha.2018.01.130
12. Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, et al. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. doi:10.1186/1476-4598-10-29
13. Feng R, Chen X, Yu Y, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298(1):50–63. doi:10.1016/j.canlet.2010.06.004
14. Wang Y, Zheng X, Zhang Z, et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7:10.
15. Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. doi:10.1186/s12943-018-0935-5
16. Shi X, Wang X, Hua Y. LncRNA GACAT1 promotes gastric cancer cell growth, invasion and migration by regulating MiR-149-mediated of ZBTB2 and SP1. J Cancer. 2018;9(20):3715–3722. doi:10.7150/jca.27546
17. Xu C, Shao Y, Xia T, et al. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol. 2014;35(10):9701–9706. doi:10.1007/s13277-014-2274-5
18. Sheng N, Xu YZ, Xi QH, et al. Overexpression of KIF2A is suppressed by miR-206 and associated with poor prognosis in ovarian cancer. Cell Physiol Biochem. 2018;50(3):810–822. doi:10.1159/000494467
19. Zhang X, Ma C, Wang Q, et al. Role of KIF2A in the progression and metastasis of human glioma. Mol Med Rep. 2016;13(2):1781–1787. doi:10.3892/mmr.2015.4700
20. Fan X, Wang X, Zhu H, Wang W, Zhang S, Wang Z. KIF2A overexpression and its association with clinicopathologic characteristics and unfavorable prognosis in colorectal cancer. Tumour Biol. 2015;36(11):8895–8902. doi:10.1007/s13277-015-3603-z
21. Zhang S, Huang F, Wang Y, Song Q, Yang X, Wu H. KIF2A overexpression and its association with clinicopathologic characteristics and poor prognoses in patients with gastric cancer. Dis Markers. 2016;2016.
22. Zhao Y, Liu Y, Lin L, et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17(1):69. doi:10.1186/s12943-018-0820-2
23. Huang Y, Zhang J, Hou L, et al. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2017;36(1):194. doi:10.1186/s13046-017-0666-2
24. Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76(21):6299–6310. doi:10.1158/0008-5472.CAN-16-0356
25. Xu MD, Wang Y, Weng W, et al. A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin Cancer Res. 2017;23(8):2071–2080. doi:10.1158/1078-0432.CCR-16-0742
26. Zhang Y, Yang R, Lian J, Xu H. LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res. 2016;8(11):5035–5043.
27. Yao XM, Tang JH, Zhu H, Jing Y. High expression of LncRNA CASC15 is a risk factor for gastric cancer prognosis and promote the proliferation of gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21(24):5661–5667. doi:10.26355/eurrev_201712_14010
28. Peng W, Wu J, Fan H, Lu J, Feng J. LncRNA EGOT promotes tumorigenesis via hedgehog pathway in gastric cancer. Pathol Oncol Res. 2019;25(3):883–887. doi:10.1007/s12253-017-0367-3
29. Chen X, Dai M, Zhu H, et al. Evaluation on the diagnostic and prognostic values of long non-coding RNA BLACAT1 in common types of human cancer. Mol Cancer. 2017;16(1):160. doi:10.1186/s12943-017-0728-2
30. Li X, Qi S, Ma D, Fan J, Wang J. Long non-coding RNA BLACAT1 promotes the proliferation and invasion of glioma cells via Wnt/beta-catenin signaling. Exp Ther Med. 2019;17(6):4703–4708. doi:10.3892/etm.2019.7468
31. Yang H, Qi Y, Wang XL, Gu JJ, Shi TM. Down-regulation of lncRNA BLACAT1 inhibits ovarian cancer progression by suppressing the Wnt/beta-catenin signaling pathway via regulating miR-519d-3p. Mol Cell Biochem. 2020;467(1–2):95–105. doi:10.1007/s11010-020-03704-y
32. Jin L, Li Y, Liu J, et al. Tumor suppressor miR-149-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Mol Med Rep. 2016;13(6):5386–5392. doi:10.3892/mmr.2016.5205
33. Chen W, Zhang J, Xu H, Dai J, Zhang X. The negative regulation of miR-149-5p in melanoma cell survival and apoptosis by targeting LRIG2. Am J Transl Res. 2017;9(9):4331–4340.
34. Li X, Liang J, Liu YX, et al. miR-149 reverses cisplatin resistance of gastric cancer SGC7901/DDP cells by targeting FoxM1. Pharmazie. 2016;71(11):640–643. doi:10.1691/ph.2016.6696
35. Zhao L, Han T, Li Y, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31(3):893–903. doi:10.1096/fj.201600994R
36. Shao Y, Ye M, Li Q, et al. LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7(25):37812–37824. doi:10.18632/oncotarget.9336
37. Peng Y, Leng W, Duan S, Hong M. Long noncoding RNA BLACAT1 is overexpressed in hepatocellular carcinoma and its downregulation suppressed cancer cell development through endogenously competing against hsa-miR-485-5p. Biomed Pharmacother. 2019;116:109027. doi:10.1016/j.biopha.2019.109027
38. Ye JR, Liu L, Zheng F. Long noncoding RNA bladder cancer associated transcript 1 promotes the proliferation, migration, and invasion of nonsmall cell lung cancer through sponging miR-144. DNA Cell Biol. 2017;36(10):845–852. doi:10.1089/dna.2017.3854
39. Liu N, Hu G, Wang H, Wang Y, Guo Z. LncRNA BLACAT1 regulates VASP expression via binding to miR-605-3p and promotes glioma development. J Cell Physiol. 2019;234(12):22144–22152. doi:10.1002/jcp.28778
40. Hu X, Liu Y, Du Y, Cheng T, Xia W. Long non-coding RNA BLACAT1 promotes breast cancer cell proliferation and metastasis by miR-150-5p/CCR2. Cell Biosci. 2019;9:14. doi:10.1186/s13578-019-0274-2
41. Wang CQ, Li YJ, Wei ZM, et al. Stable gene-silence of Kif2a synergistic with 5-fluorouracil suppresses oral tongue squamous cell carcinoma growth in vitro and in vivo. Oral Surg Oral Med Oral Pathol Oral Radiol. 2013;116(1):49–54. doi:10.1016/j.oooo.2013.01.028
42. Zhang Y, You X, Liu H, et al. High KIF2A expression predicts unfavorable prognosis in diffuse large B cell lymphoma. Ann Hematol. 2017;96(9):1485–1491. doi:10.1007/s00277-017-3047-1
43. Zhao P, Lan F, Zhang H, Zeng G, Liu D. Down-regulation of KIF2A inhibits gastric cancer cell invasion via suppressing MT1-MMP. Clin Exp Pharmacol Physiol. 2018;45(10):1010–1018. doi:10.1111/1440-1681.12974
44. Wang J, Ma S, Ma R, et al. KIF2A silencing inhibits the proliferation and migration of breast cancer cells and correlates with unfavorable prognosis in breast cancer. BMC Cancer. 2014;14(1):461. doi:10.1186/1471-2407-14-461
45. Xie T, Li X, Ye F, et al. High KIF2A expression promotes proliferation, migration and predicts poor prognosis in lung adenocarcinoma. Biochem Biophys Res Commun. 2018;497(1):65–72. doi:10.1016/j.bbrc.2018.02.020
46. Uchida A, Seki N, Mizuno K, et al. Regulation of KIF2A by antitumor miR-451a inhibits cancer cell aggressiveness features in lung squamous cell carcinoma. Cancers (Basel). 2019;11(2). doi:10.3390/cancers11020258.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.