Back to Journals » Cancer Management and Research » Volume 12
Long Intergenic Non-Protein Coding RNA 1089 Suppresses Cell Proliferation and Metastasis in Gastric Cancer by Regulating miRNA-27a-3p/Epithelial–Mesenchymal Transition (EMT) Axis
Authors Yang F, Chen X, Li X , Chen J, Tang Y, Cai Y, Wang Y, Chen Z, Li L, Li R, Deng Z
Received 18 March 2020
Accepted for publication 11 June 2020
Published 9 July 2020 Volume 2020:12 Pages 5587—5596
DOI https://doi.org/10.2147/CMAR.S254064
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Feng Yang,1,* Xiaoting Chen,2,* Xiyao Li,3,* Jianhua Chen,1 Yuxin Tang,1 Yongchang Cai,1 Yijun Wang,1 Zhiliang Chen,1 Libo Li,1 Ruiping Li,1 Zhenwei Deng1
1Department of General Surgery, Affiliated Dongguan People’s Hospital, Southern Medical University, Dongguan 523059, People’s Republic of China; 2Department of Oncology, Zhujiang Hospital, Southern Medical University, Guangzhou 510280, People’s Republic of China; 3Department of General Surgery, The First Hospital of China Medical University, Shenyang 110001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhenwei Deng; Ruiping Li
Department of General Surgery, Affiliated Dongguan People’s Hospital, Southern Medical University, Dongguan 523059, People’s Republic of China
Email [email protected]; [email protected]
Aim: To explore the expression and biological function of long intergenic non-protein coding RNA 1089 (LINC01089) in gastric cancer (GC) progression and its underlying mechanism.
Methods: LINC01089 and microRNA-27a-3p (miR-27a-3p) expressions were detected with the quantitative real-time polymerase chain reaction (RT-qPCR). Cell proliferation, migration and invasion were evaluated by Cell Counting Kit-8 (CCK-8) and Transwell assay. Epithelial–mesenchymal transition (EMT)-related proteins were also measured by Western blot. The relationship between LINC01089 and miR-27a-3p was revealed by a bioinformatics analysis and dual-luciferase reporter assay.
Results: LINC01089 was significantly down-regulated in GC tissues, as well as GC cell lines. GC patients with lower LINC01089 expression were more likely to have poor outcomes. Overexpression of LINC01089 significantly suppressed GC cells growth, migration and invasion and forbade the EMT process. LINC01089 was directly targeted at miR-27a-3p. The transfection of miR-27a-3p mimics reversed the inhibitory effects on proliferative and metastatic abilities of GC cells with LINC01089 overexpression.
Conclusion: LINC01089 inhibits cell proliferation and metastasis in GC by targeting miR-27a-3p/EMT axis, which should be considered as a promising therapeutic target.
Keywords: LINC01089, miRNA-27a-3p, proliferation, metastasis, epithelial-mesenchymal transition (EMT), gastric cancer (GC)
Introduction
Gastric cancer (GC) ranks fifth in terms of commonly diagnosed cancer and represents the third cause of cancer-related deaths worldwide.1 For the lack of early diagnostic markers, most of the GC patients were confirmed at the locally advanced or metastasis stage.2 Radical surgery resection combined with perioperative chemotherapy remains the first line of treatment for GC, but the survival and prognosis of GC was still unsatisfying.3 Hence, it is urgent to identify novel predictive and prognostic biomarkers for GC and illuminate its molecular mechanism.
Long non-coding RNAs (lncRNAs) are defined as a group of non-coding RNAs molecules longer than 200 nucleotides. In recent years, lncRNAs have been implicated in the development of various cancer, including colorectal cancers, ovarian cancer and liver cancer, in which they might function as potential oncogenes or cancer-suppressor RNAs.4,5 Furthermore, emerging evidence proved that lncRNAs might act as a transcriptional or post-transcriptional regulator, by inducing epigenetic silencing, sponging microRNAs, or controlling RNA processing and stability.6,7 For example, lncRNA HOTAIR facilitates carcinogenesis in acute myeloid leukemia via epigenetic silencing of p15.8 MALAT1 enhances cell proliferation, migration and invasion in HCC by acting as a competing endogenous RNA (ceRNA) sponge for miR-142-3p.9 To date, numerous lncRNAs have not been clearly elucidated in cancers, whereas the involvement of LINC01089 in GC progression is our particular interest.
LINC01089, also known as LncRNA Inhibiting Metastasis (LIMT), is a non-coding RNA oriented from chromosome 12. Previously, LINC01089 was recognized as a cancer-suppressor in breast cancer. For example, Chen reported that ectopic expression of LINC01089 reduced migratory and invasive capacities of MCF10A cells.10 Similarly, Yuan et al provided evidence that LINC01089 blocked Wnt/β-catenin signaling and then suppressed cell proliferation, migration and invasion in breast cancer cells.11 Based on these studies, we hypothesized LINC01089 might be involved in GC and its potential regulatory mechanism should be investigated further.
Herein, we firstly analyze the relationship between clinicopathological characteristics and LINC01089 expression in GC patients. Next, we conducted functional experiments to identify the biological role of LINC01089 in GC cells. Finally, bioinformatics analysis and dual-luciferase reporter assays revealed that LINC01089 directly binds to miR-27a-3p, and therefore inhibited the carcinogenesis of GC. Altogether, our results reveal LINC01089 as a promising prognostic indicator and a novel therapeutic biomarker for GC patients.
Patients and Methods
Patient Tissue Samples
Our study comprised of a total of eighty pairs of GC tissue samples and their adjacent normal tissues from patients who had undergone a gastrectomy in Dongguan People’s Hospital from 2009 to 2012 and had signed the informed consent voluntarily. All samples were histologically confirmed as gastric adenocarcinoma by two independent pathologists. Sample collection was in accordance with the requirements of the Ethics Committee of Dongguan People’s Hospital and informed written consent was obtained from each patient.
Cell Culture
Gastric mucosal cells (GES-1) and GC cell lines (AGS, BGC-823, HGC-27, MGC-803, MKN45) were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). All cell lines were in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS;GIBCO, Grand Island, NY, USA) and were incubated at 37°C and with 5% CO2.
Cell Transfection
The LINC01089 full-length sequence was synthesized and subcloned into a pcDNA3.0 vector (Invitrogen). The hsa-miR-27a-3p mimics were synthesized by RiboBio Co Ltd (Guangdong, China). Based on the manufacturer’s protocol, LipofectamineTM 3000 reagent (Invitrogen) was used to transfect the LINC01089 plasmid or miR-27a-3p mimics into GC cells.
Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted with TRIzol reagent (Invitrogen) and then subjected to reverse transcription and PCR reactions using Primescript reverse transcriptase (Takara, Kusatsu, Japan) based on the manufacturer’s instructions. In the RT-qPCR assay, the expression levels of LINC01089 or miR-27a-3p were determined using PrimeScriptTM RT-PCR kit (Takara) or TaqMan MicroRNA Assays Kit (Applied Biosystems, Carlsbad, CA, USA). Relative gene expression was quantified when compared to internal control GAPDH or U6 using comparative CT method. Primer sequences were: LINC01089, Forward: 5`-TTT CCC TGC CTG CCC TCC AG-3` and Reverse: 5`-CAG TTG TGC TCC AGC CTT CCA G −3`; GAPDH Forward: 5`-GGT GTG AAC CAT GAG AAG TAT GA-3` and Reverse: 5`-GAG TCC TTC CAC GAT ACC AAA G-3`; miR-27a-3p, Forward: 5`-GCA GTT CAC AGT GGC GGC TAA G-3` and Reverse: 5`-CCA GTT TTT TTT TTT TTT TGC GGA-3`; U6 Forward: 5`-CTC GCT TCG GCA GCA CA-3` and Reverse: 5`-AAC GCT TCA CGA ATT TGC GT-3`.
Cell Growth Assay
A Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) assay was performed in our study to evaluate GC cells’ viability at the designated time interval based on the manufacturer’s instructions. OD value was determined by a micro-spectrophotometer.
Transwell Assay
Cell migration and invasion were performed using Transwell chambers (8-μm pore size; Corning, NY, USA) with or without Matrigel (BD, San Diego, CA, USA) pre-coating, respectively.
GC cells were harvested and suspended in a serum-free medium. The cell suspension was added to the upper chamber, while a medium containing 20% FBS was added to the lower chamber. The non-migrated or invaded cells were removed. The remaining cells were fixed, stained with crystal violet and then observed under a microscope.
Western Blot Assay
Ice-cold RIPA buffer (Beyotime, Shanghai, People’s Republic of China) was used to lyse the GC cell precipitations for protein extraction. Equal amounts of protein determined by a BCA Protein Assay Kit (Beyotime) were separated by SDS-PAGE and then transferred onto PVDF membranes. Ten per cent BSA solutions were incubated with the PVDF membranes to block the unspecific antibody. The membranes were incubated with the following primary antibodies: E-Cadherin, N-Cadherin, Vimentin, GAPDH. The next day they were incubated with an HRP labeled secondary antibody. After being rinsed with PBST, the protein bands were visualized using GeneSnap software on SynGene system (Cambridge, UK). The gray value was analyzed to determine the the relative expression of the target genes using GAPDH as an internal control.
Dual-Luciferase Reporter Assays
293T cells were co-transfected with pmirGLO- LINC01089 -WT or pmirGLO- LINC01089-MUT plasmid, with miR-27a-3p mimics or miR-27a-3p NC by LipofectamineTM 3000 reagent. A dual-luciferase reporter assay system (Promega, Madison, WI) was used to detect the relative luciferase activity based on the manufacturer’s protocol.
Statistical Analysis
All data were obtained from three repeated experiments and were analyzed using IBM SPSS 20.0 software. Comparisons between the two groups and multiple groups were analyzed by Student’s t-test and Chi-square test. P<0.05 was considered statistically significant.
Results
LINC01089 Is Down-Regulated in GC and Is Associated with Poor Oncologic Outcomes
As Figure 1A has shown, LINC01089 was found at a low levels in GC tissue samples, when compared with adjacent normal tissues. Meanwhile, downregulation of LINC01089 accounted for 76.3% (61/80) of GC tissue samples (Figure 1B). According to the median of the relative LINC01089 expression, the selected GC patients were divided into high expression group and low expression group. Statistical analysis indicated that LINC01089 expression was tightly associated with T stage (P=0.0039), N stage (P=0.0036), TNM stage (P=0.044) and vascular invasion (P=0.003) (Table 1). GC patients with accompanied with advanced T stage (Figure 1C, P=0.002), lymph node metastasis (Figure 1D, P=0.007), advanced TNM stage (Figure 1E, P=0.029), presence of vascular invasion (Figure 1F, P=0.009) or shorter overall survival (Figure 1G, P=0.022) exhibited considerably lower LINC01089 expression. Taken together, low expression of LINC01089 was identified in GC and was closely related to poor clinical outcomes of GC patients.
 |
Table 1 Correlation Between LINC01089 Expression and Clinicopathological Features in Patients with Gastric Cancer (GC) |
Overexpression of LINC01089 Suppresses GC Cells Proliferation
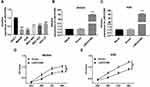
Following the evidence that LINC01089 was downregulated in GC tissues, we next validated its biological impact in GC progression. Initially, the expression levels of LINC01089 was detect in a cohort of GC cell lines. Gastric mucosal cells was set as a control. The results showed that LINC01089 expression was down-regulated in GC cell lines compared with GES-1 cell lines (Figure 2A). Given that AGS and MNK-45 cell lines showed the lower LINC01089 expression, we then transfected an overexpression plasmid or an empty vector into these cells. RT-qPCR confirmed that LINC01089 was successfully overexpressed in AGS and MNK-45 cell lines with overexpression plasmid compared to the cells with empty vector (Figure 2B and C). CCK8 assay was performed to evaluate cell proliferation. The growth curve indicated that the growth ability of AGS and MNK-45 cells with LINC01089 overexpression was significantly weakened compared with the control cells (Figure 2D and E), indicating that overexpression of LINC01089 suppressed cell proliferation in GC.
Overexpression of LINC01089 Inhibits GC Cells Migration and Invasion and Suspends Epithelial–Mesenchymal Transition Process in GC
Our statistical analysis showed that GC patients with lower expression of LINC01089 are prone to have local lymph node metastasis and vascular invasion, which might contribute to the enhancement of GC cells metastasis. Therefore, a Transwell assay was performed to detect cell migration and invasion. Our data observed that increased AGS and MNK-45 cells were obvious in the LINC01089 overexpression group when compared to the empty vector group (Figure 3A–D), indicating that overexpression of LINC01089 inhibits cell migration and invasion in GC.
Upon establishing the effect of LINC01089 in the regulation of GC cell migration and invasion, we investigated the underlying mechanism. Considering that the development of invasive and metastatic cancer always follows an epithelial-mesenchymal transition (EMT) process, we assessed the EMT phenotypic markers in GC cells by Western blot assay. The result revealed that overexpression of LINC01089 in AGS and MNK-45 cells increased the expression of epithelial-like phenotype (E-cadherin), accompanied by the reduction of mesenchymal phenotype (N-cadherin, Vimentin) (Figure 3E–H). These data suggested that LINC01089 might inhibit cell proliferation and metastasis by regulating the EMT process in GC.
miR-27a-3p Is a Direct Target of LINC01089 in GC
Accumulated evidence has indicated that lncRNAs exerted biologic effects by interacting with miRNAs. To determine the molecular mechanism by which LINC01089 suppresses cell proliferation and metastasis in GC, we firstly acquired its underlying target miRNAs, using the Starbase.2. We predicted that miR-27a-3p might be a downstream target for LINC01089. Expression analysis showed that miR-27a-3p was dramatically up-regulated in GC tissue samples relative to normal samples (Figure 4A), and overexpression of miR-27a-3p accounted for 85% (68/80) of GC specimens (Figure 4B). Additionally, miR-27a-3p expression was elevated in the five GC cell lines (Figure 4C). A correlation analysis showed that the relative expression of miR-27a-3p was reversely correlated with LINC01089 in GC (Figure 4D). To verify the direct interaction between LINC01089 and miR-27a-3, we predicted their potential binding site (Figure 4E), and performed dual-luciferase reporter assays, in which we observed that only the miR-27a-3p mimics could repress the LINC01089-wild type-driven luciferase activity, but not the negative control mimic or the LINC01089-mutant-driven luciferase activity (Figure 4F). Our data suggested that miR-27a-3p might be a direct downstream target of LINC01089 in GC.
miR-27a-3p Is Critical for the Function of LINC01089 in GC
To study whether miR-27a-3p mediates the anti-tumor function of LINC01089 in GC cells, we co-transfected the LINC01089 overexpression plasmid with miR-27a-3p mimics into the MNK-45 cells. The miR-27a-3p expression was confirmed by RT-qPCR assay (Figure 5A). We learned that miR-27a-3p expression was decreased in LINC01089 overexpressed MNK-45 cells, which could recover to a certain extent following the transfection of miR-27a-3p mimics. CCK-8 and Transwell assays demonstrated that LINC01089 overexpression-mediated inhibition of the cell growth and metastasis was partially rescued by co-transfection with the miR-27a-3p mimics (Figure 5B–D). A similar effect was also shown in Western blot detection on EMT-related markers. Increased E-cadherin expression and reduced N-cadherin and Vimentin expression was found in cells with co-transfection of LINC01089 overexpression plasmid with miR-27a-3p mimics, compared with the control cells (Figure 5E and F). Taken together, these results suggested that LINC01089 inhibited cell growth and metastasis occured by targeting miR-27a-3p in GC.
Discussion
In recent years, high-throughput sequencing and bioinformatics technologies have made a great breakthrough. Mounting evidence showed that lncRNAs are identified as critical regulators in GC and their pathogenetic roles were widely investigated.12 In our study, we firstly report the aberrant expression of LINC01089 in GC. The RT-qPCR assay revealed that the expression levels of LINC01089 in the GC tissues were much lower than that in the adjacent normal tissues. A similar expression pattern was also observed in GC cell lines. Further statistical analysis indicated that patients with reduced LINC01089 expression exhibited malignant features and a shorter overall survival time, which is in agreement with previous findings for breast cancer and glioma.11,13 This evidence suggested that LINC01089 should be considered as a negative prognostic factor for cancers. Accordingly, we can preliminarily conclude that LINC01089 downregulation is associated with disease progression in GC and predicts an unfavorable outcome for GC patients.
Regarding that LINC01089 might act as a cancer-suppressor in GC, we introduced two GC cell lines with co-transfection with a LINC01089 overexpression plasma to study the exact functions of LINC01089 in this disease. Patients with advanced tumor stage and lymphatic metastasis were linked to lower LINC01089 expression, indicating the important role of LINC01089 involved in cancer growth and metastasis. Generally, cell growth and metastatic capacity functioned as an important indicator to determine the malignant feature of cancer. Proliferation and metastasis deficiency have been revealed in breast cancer cells following LINC01089 overexpression.11 In CCK-8 and Transwell assay, we also observed that LINC01089 overexpression led to marked repression on cell proliferation, migration and invasion in GC.
LINC01089 was also related to blood vascular invasion, which was revealed by our clinicopathological features analysis in a GC tissue cohort. Accumulated studies demonstrated that cancer cells escaped from primary location by transforming their phenotypic characteristics so that they could invade the vascular system.14 The changes were attributed to a key subsequence of EMT, which plays an essential role in cancer cell proliferation and metastasis abilities.15 LncRNAs have merged as positive or negative players in the regulation and induction of EMT in GC.16,18 For example, Zhou et al reported that SNHG16 promotes EMT via down-regulation of DKK3 in GC cells.19 Conversely, Wu et al demonstrated that HRCEG levels inversely correlate with EMT process by interacting with HDAC1 in GC.20 EMT enhances the metastasic potential of epithelial cells by the loss of their epithelial phenotype and the gain of mesenchymal characteristics.21,22 To further illuminate the regulatory role of LINC01089 in GC metastasis, we conducted a Western blot assay to determine the change of the EMT related marker. The results showed that apart from down-regulating Vimentin and N-cadherin, LINC01089 overexpression also increased E-cadherin expression, suggesting that LINC01089 is important for restraining EMT progression in GC.
Due to the cavernous mechanism, lncRNAs could directly impair the function of miRNAs, and therefore exert cancer suppression or cancer promotion effect.23,25 For example, Ren et al demonstrated that DDX11-AS1 may serve as an oncogene in GC progression by interplaying with miR-873-5p.26 Dong et al reported that the silencing of PCAT6 inhibits proliferation and EMT of GC cells by targeting miR-15a.27 Our bioinformatics analysis and luciferase reporter assays revealed that miR-27a-3p is a target of LINC01089. In addition, the expression of miR-27a-3p was enhanced in GC cells upon LINC01089 overexpression. Moreover, a negative relation between LINC01089 and miR-27a-3p was also found in the GC sample cohort. Notably, miR-27a-3p has been reported as an oncogenic non-coding RNA in GC by promoting GC cell proliferation in vitro as well as tumor growth in vivo.28 In another study in GC, miR-27a-3p was revealed to promote EMT by targeting NOVA alternative splicing regulator 1.29 In the present study, the introduction of miR-27a-3p mimics in GC cells reversed the inhibitory effect on cell proliferation, metastasis, and EMT process induced by LINC01089 overexpression. Taken together, our findings demonstrated the significance of the interaction between LINC01089 and miR-27a-3p in GC carcinogenesis, given that LINC01089 exerts a cancer-suppressing function partly via targeting miR-27a-3p in GC cells.
In conclusion, We have identified a novel cancer suppressor gene in the GC procession. We demonstrated LINC01089 was downregulated in GC tissues and contributed to GC progression. The key findings in our study demonstrate that LINC01089 could directly regulate the function of miR-27a-3p/EMT axis, which represents a mechanism by which LINC01089 inhibits cell proliferation, migration and invasion in GC. Collectively, we propose that LINC01089 targeting therapies underlines a beneficial way to treatGC patients.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Khanderia E, Markar SR, Acharya A, Kim Y, Kim YW, Hanna GB. The influence of gastric cancer screening on the stage at diagnosis and survival: a meta-analysis of comparative studies in the Far East. J Clin Gastroenterol. 2016;50(3):190–197. doi:10.1097/MCG.0000000000000466
3. Brenkman HJ, Haverkamp L, Ruurda JP, van Hillegersberg R. Worldwide practice in gastric cancer surgery. World J Gastroenterol. 2016;22(15):4041–4048. doi:10.3748/wjg.v22.i15.4041
4. Dai X, Kaushik AC, Zhang J. The emerging role of major regulatory RNAs in cancer control. Front Oncol. 2019;9:920. doi:10.3389/fonc.2019.00920
5. Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–1967. doi:10.1002/ijc.30546
6. Li D, Yang C, Yin C, et al. LncRNA, important player in bone development and disease. Endocr Metab Immune Disord Drug Targets. 2020;20(1):50–66. doi:10.2174/1871530319666190904161707
7. Hao NB, He YF, Li XQ, Wang K, Wang RL. The role of miRNA and lncRNA in gastric cancer. Oncotarget. 2017;8(46):81572–81582. doi:10.18632/oncotarget.19197
8. Gao S, Zhou B, Li H, et al. Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Exp Hematol. 2018;67:32–40 e33. doi:10.1016/j.exphem.2018.08.005
9. Yu Q, Xiang L, Chen Z, et al. MALAT1 functions as a competing endogenous RNA to regulate SMAD5 expression by acting as a sponge for miR-142-3p in hepatocellular carcinoma. Cell Biosci. 2019;9:39. doi:10.1186/s13578-019-0299-6
10. Sas-Chen A, Aure MR, Leibovich L, et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol Med. 2016;8(9):1052–1064. doi:10.15252/emmm.201606198
11. Yuan H, Qin Y, Zeng B, et al. Long noncoding RNA LINC01089 predicts clinical prognosis and inhibits cell proliferation and invasion through the Wnt/beta-catenin signaling pathway in breast cancer. Onco Targets Ther. 2019;12:4883–4895. doi:10.2147/OTT.S208830
12. Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142(11):2231–2237. doi:10.1007/s00432-016-2183-7
13. Hassan A, Mosley J, Singh S, Zinn PO, Comprehensive A. Review of genomics and noncoding RNA in gliomas. Top Magn Reson Imaging. 2017;26(1):3–14. doi:10.1097/RMR.0000000000000111
14. Li P, Ling YH, Zhu CM, et al. Vascular invasion as an independent predictor of poor prognosis in nonmetastatic gastric cancer after curative resection. Int J Clin Exp Pathol. 2015;8(4):3910–3918.
15. Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. doi:10.1038/nrc.2017.118
16. Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139(2):269–280. doi:10.1002/ijc.30039
17. Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205–217. doi:10.3727/096504016X14549667334007
18. Wan X, Ding X, Chen S, et al. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumour Biol. 2015;36(2):521–532. doi:10.1007/s13277-015-3136-5
19. Zhou C, Zhao J, Liu J, et al. LncRNA SNHG16 promotes epithelial- mesenchymal transition via down-regulation of DKK3 in gastric cancer. Cancer Biomark. 2019;26(4):393–401. doi:10.3233/CBM-190497
20. Wu S, Wu E, Wang D, et al. LncRNA HRCEG, regulated by HDAC1, inhibits cells proliferation and epithelial-mesenchymal-transition in gastric cancer. Cancer Genet. 2020;241:25–33. doi:10.1016/j.cancergen.2019.12.007
21. De Wever O, Pauwels P, De Craene B, et al. Molecular and pathological signatures of epithelial-mesenchymal transitions at the cancer invasion front. Histochem Cell Biol. 2008;130(3):481–494. doi:10.1007/s00418-008-0464-1
22. Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17(5):548–558. doi:10.1016/j.ceb.2005.08.001
23. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. doi:10.1016/j.bbagrm.2015.06.015
24. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi:10.1016/j.bbagrm.2014.08.012
25. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi:10.1016/j.semcdb.2014.05.015
26. Ren Z, Liu X, Si Y, Yang D. Long non-coding RNA DDX11-AS1 facilitates gastric cancer progression by regulating miR-873-5p/SPC18 axis. Artif Cells Nanomed Biotechnol. 2020;48(1):572–583. doi:10.1080/21691401.2020.1726937
27. Dong D, Lun Y, Sun B, et al. Silencing of long non-coding RNA PCAT6 restrains gastric cancer cell proliferation and epithelial-mesenchymal transition by targeting microRNA-15a. Gen Physiol Biophys. 2020;39(1):1–12. doi:10.4149/gpb_2019044
28. Zhou L, Liang X, Zhang L, et al. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget. 2016;7(32):51943–51954. doi:10.18632/oncotarget.10460
29. Li K, Zhu X, Chen X, Wang X. MicroRNA27a3p promotes epithelialmesenchymal transition by targeting NOVA alternative splicing regulator 1 in gastric cancer. Mol Med Rep. 2020;21(3):1615–1622. doi:10.3892/mmr.2020.10949
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.