Back to Journals » OncoTargets and Therapy » Volume 13
Long-Chain Non-Coding RNA SNHG3 Promotes the Growth of Ovarian Cancer Cells by Targeting miR-339-5p/TRPC3 Axis
Authors Liu EL, Zhou YX, Li J, Zhang DH, Liang F
Received 14 February 2020
Accepted for publication 31 August 2020
Published 28 October 2020 Volume 2020:13 Pages 10959—10971
DOI https://doi.org/10.2147/OTT.S249873
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
En-Ling Liu,1 Yu-Xiu Zhou,2 Jun Li,1 Dong-Hong Zhang,1 Feng Liang1
1The Department of Obstetrics and Gynecology, Tangshan Gongren Hospital Affiliated to Hebei Medical University, Tangshan, Hebei Province, People’s Republic of China; 2The Department of Immunology, Tangshan Gongren Hospital Affiliated to Hebei Medical University, Tangshan, Hebei Province, People’s Republic of China
Correspondence: En-Ling Liu
The Department of Obstetrics and Gynecology, Tangshan Gongren Hospital Affiliated to Hebei Medical University, No. 27 Wenhua Road, Lubei District, Tangshan, Hebei Province, People’s Republic of China
Tel +0315-3722602
Email [email protected]
Background: Long-chain non-coding RNA (lncRNA) small nucleolar RNA host gene 3 (SNHG3) is reportedly overexpressed in malignant tumors, but its regulatory role in human ovarian cancer (OC) is not fully understood.
Methods: A qRT-PCR assay was carried out to detect the level of SNHG3 in OC tissues, serum and cells, a CCK-8 assay to measure the proliferation of OC cells, a transwell assay to measure the invasion and migration of OC cells, and a flow cytometry to detect the cell cycle distribution and apoptosis rate of OC cells. In addition, in vivo experiment was also conducted to determine the effect of SNHG3 on the growth of OC cells.
Results: SNHG3 was overexpressed in OC tissues, serum, and cells, and the overexpression in serum indicated a poor prognosis of patients. It was also found that knockdown of SNHG3 could inhibit the malignant phenotypes of OC cells, cause G1/G0 cell cycle arrest, and intensify apoptosis. Furthermore, in in vitro experiments, the growth ability of OC cells was inhibited under knockdown of SNHG3. Assays for relationship verification showed that SNHG3 regulated the expression of miR-339-5p and the canonical transient receptor potential 3 (TRPC3), and the rescue experiment revealed that co-transfection of si-SNHG3+miR-339-5p-inhibitor or si-SNHG3+pcDNA3.1-TRPC3 could reverse the effects of knockdown of SNHG3 on the biological behavior of OC cells.
Conclusion: SNHG3 can be adopted as a marker for diagnosis and prognosis evaluation of OC and it plays a role in the progression of OC by enabling the miR-339-5p sponge to regulate TRPC3 expression.
Keywords: long-chain non-coding RNA, small nucleolar RNA host gene 3, miR-339-5p, ovarian cancer, biological behavior
Introduction
Ovarian cancer (OC) is a prevalent malignant tumor of female genital organs in gynecology, with a high incidence.1 It usually occurs in the deep pelvic cavity with a small tumor focus and lacks clinical symptoms at early stage. At this point, there is no reliable test to diagnose ovarian cancer before patients have symptoms, so nearly 70% of the patients are already at the advanced stage at the time of diagnosis, and the 5-year survival rate of patients with ovarian cancer is lower than 30%.2 Clinically, ovarian neoplasms are identified by testing markers in blood or by ultrasound images, but these tests are usually used too late for patients, and sometimes they will produce “false positive” results, which may cause unnecessary surgery for women without cancer.3 Despite the upgrading of surgical treatment and chemotherapy, the prognosis of patients with OC is still not optimistic.4 Therefore, understanding the mechanism of OC may provide insights for early diagnosis and personalized treatment of OC.
There are evidences that the onset of OC is a highly complicated process involving changes in epigenetic mechanisms.5 Long-chain non-coding RNA (lncRNA) is a regulator of gene expression involving the development of various diseases, and its imbalance can often be found in the serum or tissues of malignant tumors.6,7 The imbalance of lncRNA expression is linked to the progress of malignant tumors, including OC, and it can mediate the malignant behavior of cancer cells, thus regulating the process of cancer metastasis and differentiation.8 The role of lncRNA in OC has long captured extensive attention.9,10 Small nucleolar RNA host gene 3 (SNHG3), an lncRNA, acts as a carcinogen in various tumors.11,12 For example, it is overexpressed in hepatic carcinoma and is strongly linked to malignant recurrence and prognosis of patients.13 There is also a report from Li et al14 that SNHG3 is not only related to the prognosis of patients with OC, but also related to the energy metabolism of patients. Although there are many reports on SNHG3 in various malignant tumors including OC, there is little research on the clinical effect and molecular mechanism of it in OC. One previous study has found that SNHG3, up-regulated in osteosarcoma, has potential in the diagnosis and prognosis of patients with osteosarcoma, and can regulate cell invasion and migration by mediating RAB22A via miR-151a-3p.15 We found through an online biological database that miR-339-5p is linked to SNHG3, but the role of the two in OC has not been studied.
In this study, we explored the role and molecular mechanism of SNHG3 in OC through experiments.
Materials and Methods
Sample Collection
Peripheral blood, tumor tissues, and corresponding tumor-adjacent tissues were sampled from 96 patients with OC from March 2011 to April 2014. The patients were confirmed with OC according to histopathology.16 The patients had not received any anti-tumor treatment before surgery, and each of them had an expected survival time longer than 1 month. In addition, they were not complicated with other malignant tumors. Peripheral blood was also sampled from 96 healthy individuals in physical examination during the same period. Elbow venous blood (5 mL) was sampled and centrifuged at 4°C and 800g for 10 min, and then the supernatant was transferred to test tubes, and stored for later analysis. Each subject signed an informed consent form after understanding this study, and the study was carried out with the permission of the Ethics Committee of Tangshan Gongren Hospital Affiliated to Hebei Medical University.
Cell Culture
Human ovarian cancer cell strains (SKOV3, HeyA8, and A2780) and immortalized human non-malignant ovarian surface epithelium (IOSE) cells purchased from Shun Ran Biological Technology Co., Ltd. (Shanghai, China) were cultured in RPMI-1640 medium (Haoran Bio Technologies Co., Ltd., Shanghai, China) supplemented with 10% fetal bovine serum (FBS) + 100 U/mL penicillin/streptomycin at 37 °C under 5% CO2.
qRT-PCR Assay
Total RNA was extracted using a TRIzol Kit (Invitrogen) under the kit instructions, and the RNA was quantified and purified using a UV ultraviolet photometer (Aoyi Instruments Co., Ltd., Shanghai, China). Total RNA reactant was reversely transcribed into cDNA with PrimeScript reagent (Applied Biosystems), and PCR quantification was carried out to it on an AIB 7500 real-time PCR system (Thermo Fisher Scientific., Shanghai, China) through a PrimeScript RT Master Mix Kit three times. GADPH was taken as an internal reference for mRNA, and U6 for miR in the experiment, and the relative expression of mRNA and miR was calculated through 2-ΔΔCT.17 The sequences were designed by Thermo Fisher Scientific (Shanghai, China), and they are as follows: for SNHG3, forward: 5ʹ-TTCAAGCGATTCTCGTGCC-3ʹ, reverse: 5ʹ-AAGATTGTCAAACCCTCCCTGT-3ʹ; for miR-339-5p, forward: 5ʹ-GGGTCCCTGTCCTCCA-3ʹ, reverse: 5ʹ-TGCGTGTCGTGGAGTC-3ʹ; for GAPDH: forward: 5ʹ-TATGATGATATCAAGAGGGTAGT-3ʹ, reverse: 5ʹ-TGTATCCAAACTCATTGTCATAC-3ʹ; for U6, forward: 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ, reverse: 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ.
Cell Transfection
The sequence of full-length SNHG3 or the canonical transient receptor potential 3 (TRPC3) was imported into pcDNA3.1 vectors (Bei Nuo Biotechnology Co., Ltd., Shanghai, China) to realize overexpression of SNHG3 (pcDNA3.1-SNHG3) and TRPC3 (pcDNA3.1-TRPC3), and blank vectors were used as controls. Small interfering SNHG3 (si-SNHG3), miR-339-5p mimics, and miR-339-5p inhibitor were purchased from Thermo Fisher Scientific (Shanghai, China). Plasmids or siRNA oligonucleotide were transfected into SKOV3 and HeyA8 cells by Lipofectamine 2000 reagent (Bei Nuo Biotechnology Co., Ltd., Shanghai, China) after the cells were cultured overnight at 5× 104 cells/well.
Cell Proliferation Assay
CCK8 was adopted to measure cell proliferation. The cells were transferred to a 96-well plate at 1×103 cells per well, and then incubated in a 5% CO2 cell incubator at 37°C. CCK-8 solution (10 μL) was added into the cells at a specific time, and the cells were cultured continually for 2 h after addition of the solution each time. After the culturing, the optical density (OD) of each well at 450 nm was determined with a microplate reader (Molecular Devices, Shanghai, China).
Transwell Assay
A transwell assay was carried out to detect cell invasion and migration. In the cell migration assay, the cells were added into the upper compartment at 5×104 cells per well, while in the cell invasion assay, the cells were added into the upper compartment at 1× 105 cells per well. The matrix gelatin was laid, and 200 μL cell suspension was transferred to the upper compartment, and 600 μL medium with 10% FBS was transferred to the lower compartment. The whole chamber was incubated in an incubator. After 24 hours, the cells or matrix gelatin that did not penetrate the membrane in the upper compartment were wiped off using cotton swabs, and the remaining cells were immobilized with 1% formaldehyde, followed by 0.1% crystal violet dyeing for 15min. Finally, the number of cells that had penetrated the membrane in five randomly selected fields under a microscope was calculated.
Apoptosis and Protein Cycle
Cells were collected after transfection, and their apoptosis was detected with an Annexin V-FITC Apoptosis Detection Kit (Bei Nuo Biotechnology Co., Ltd., Shanghai, China) under kit instructions. The collected cells were stained with propidium iodide (100 μg/mL), ribonuclease A (10 μg/mL), and phosphate buffer saline (PBS) (290 μL) in the dark for 30 min, followed by a filtering by a 300 mesh screen. Afterwards, apoptosis and cell cycle of the cells were evaluated using a flow cytometer (IMAGE Trading Co., Ltd., Beijing, China).
Luciferase Reporter Gene Assay
The potential targets of SNHG3 and miR-339-5p were predicted by TargetScan online database or starBase v3.0.18 Wild-type (WT) SNHG3, TRPC3 3ʹ-UTR, and mutant-type (MUT) SNHG3, TRPC3 3ʹ-UTR sequences were imported into psiCHEK2.0 vectors (Thermo Fisher Scientific, Shanghai, China) to construct TRPC3-WT, SNHG3-WT, TRPC3-MUT and SNHG3-MUT plasmids. WT or MUT SNHG3 3ʹ-UTR, TRPC3 3ʹ-UTR, and miR-339-5p-mimics were co-transfected into HeyA8 cells by Lipofectamine 2000 reagent. After 24 h, the cells were obtained, and their luciferase activity was detected with a Dual-Luciferase Reporter Gene Assay Kit (Qunji Biotech Co., Ltd., Shanghai, China).
RNA Immunoprecipitation (RIP)
RIP analysis was carried out according to the Magna RIP™ RNA Immunoprecipitation Kit (Thermo Fisher Scientific, Shanghai, China) instructions. When the confluency of cells reached 80%-90%, the cells were lysed in RIP lysis buffer, and the whole-cell extract was cultured with magnetic beads coupled with mouse immunoglobulin G (IgG), human anti-Ago2 antibody (XinYu Biological Technology Co., Ltd., Shanghai, China), and controls. The co-precipitated RNA was determined using a qRT-PCR assay.
RNA Pull-Down Assay
Biotinylated miR-339-5p-WT, miR-339-5p-MUT, and biotinylated NC (Thermo Fisher Scientific, Shanghai, China) were transfected into HeyA8 cells by Lipofectamine 2000 reagent, respectively. The concentration of each biotinylated miRNA was 20 nM. After 48 hours of transfection, cell lysates were harvested and cultured with dynabeads M-280 streptavidin (Invitrogen, USA), and the RNA separated by TRIzol reagent was determined using a qRT-PCR assay.
Nude Mouse Tumorigenesis Test
With five-week-old female BALB/c nude mice (n=14) under pathogen-free conditions, the experiment was carried out according to the regulations of the Animal Committee of our hospital with the permission of the Ethical Committee of Tangshan Gongren Hospital Affiliated to Hebei Medical University. HeyA8 cells stably transfected with si-NC and si-SNHG3 were obtained, and 2×107 cells were suspended into PBS solution. Suspended cells (0.1 mL) were transfected into any posterior side of each nude mouse, and when the implant became larger, the tumor volume and weight were metered once every 7 days. After 7 weeks, the mice were killed to take their tumor tissues for mass measurement. All the experiments involving animals in the study were approved by the Animal Ethics Committee of Tangshan Gongren Hospital Affiliated to Hebei Medical University and carried out under the guidance of the Animal Ethics Committee and in line with the institutional ethics guidelines for animal experiments.
Western Blotting
The cells (about 1×107 cells) were lysed using RIPA lysis buffer (Invitrogen, USA) to extract protein, and the extracted protein was quantified using a Bicinchoninic Acid (BCA) kit, and then separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated protein was sealed with 5% skimmed milk for 2 h, and washed three times with tris-buffered saline with tween 20 (TBST) buffer, 5 min each time, and the membrane was then cultured with primary anti-TRPC3 (1:1000) and internal reference of β-actin (1: 1000) at 4°C for one night. Afterwards, the membrane was washed again three times with TBST buffer, 5 min each time, and cultured with goat anti-mouse/rabbit IgG (1:1000; Invitrogen, USA) secondary antibody coupled with horseradish peroxidase at room temperature for 2 h. Finally, the membrane was washed three times with TBST buffer, 5 min each time, and developed with electrochemiluminescence (ECL) solution. The bands were analyzed using ECL (Invitrogen, USA). The assay was repeated three times.
Statistical Analysis
The data are expressed as the mean ± standard deviation (Mean±SD), and comparison of statistical values was carried out using the Student’s t test. Enumeration data are expressed as a percentage (%) and the chi-square test is expressed as χ2. Receiver operating characteristic (ROC) curves were drawn to analyze the diagnostic ability of SNHG3, and the area under the curve (AUC) of it was calculated. In addition, Kaplan–Meier curves were also drawn to analyze the overall survival rate, and Log rank test was adopted to evaluate the difference of overall survival. Pearson correlation analysis was carried out to analyze correlation. P<0.05 implies a significant difference.
Results
SNHG3 is Overexpressed in OC and the Overexpression is Related to a Poor Prognosis of Patients with OC
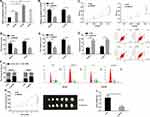
The qRT-PCR assay revealed that the SNHG3 expression in OC tissues was higher than that in corresponding tumor-adjacent tissues (Figure 1A), and it also revealed that the serum SNHG3 expression in patients with OC was significantly higher than that in healthy individuals, and the AUC of it for diagnosis of OC was 0.863 (Figure 1B). Additionally, the expression of serum SNHG3 in patients with OC was positively correlated with that in the tissues of them (Figure 1C), and the high expression of serum SNHG3 was related to lymph node metastasis and histological grading of patients with OC (Table 1 and Figure 1D–F). It is noteworthy that patients with OC with high expression of SNHG3 had a poorer prognosis than those with low expression of it (Figure 1G). The Cox proportional hazards analysis revealed that high SNHG3 expression, lymph node metastasis, and histological grade were prognostic factors for patients with OC (Table 2). The results suggested that the up-regulating SNHG3 may be linked to the occurrence, development and prognosis of patients with OC.
 |
Table 1 Relationship Between SNHG3 Expression and Clinicopathological Features of Patients with OC (n=96) |
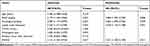 |
Table 2 Univariate and Multivariate Regression Analyses of Relevant Prognostic Parameters of Patients with OC |
Inhibition of SNHG3 Can Inhibit OC Cell Growth
For the purpose of studying the function of SNHG3 in OC, we quantified SNHG3 in SKOV3, HeyA8, A2780, and IOSE cells. It came out that the expression of SNHG3 in OC cells was generally higher than that in IOSE cells (Figure 2A). We selected SKOV3 and HeyA8 cells for follow-up experiments. The qRT-PCR assay revealed that SKOV3 and HeyA8 cells transfected with si-SNHG3 showed significantly lower SNHG3 expression than those transfected with si-NC (Figure 2B), and the CCK-8 assay showed that inhibition of SNHG3 significantly hindered the proliferation of SKOV3 and HeyA8 cells (Figure 2C). Additionally, the transwell assay revealed that knockdown of SNHG3 could inhibit cell invasion (Figure 2D) and migration (Figure 2E), and the flow cytometry analysis showed that compared with si-NC (Figure 2F), inhibition of SNHG3 could give rise to G1/G0 phase arrest of SKOV3 and HeyA8 cells and increase the apoptosis rate of them (Figure 2G). We carried out an in vivo experiment in nude mice to further analyze whether SNHG3 can affect the development of OC. It came out that after knock-down of SKOV3, the tumor growth (Figure 2H) and average tumor weight decreased significantly (Figure 2I), which implied that inhibition of SNHG3 expression can inhibit the growth of OC cells.
MiR-339-5p is the Inhibitory Target of SNHG3
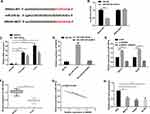
For the purpose of understanding the mechanism of SNHG3 in OC, we predicted miRNA target sites (Figure 3A) through the online database starBase3.0. The results showed that SNHG3 may be the functional target of miR-339-5p (Figure 3A). The dual-luciferase reporter assay showed that co-transfection of miR-339-5p-mimics and SNHG3-WT significantly lowered the luciferase activity of cells, while co-transfection of miR-339-5p-mimics and SNHG3-MUT exerted no effect on that of cells (Figure 3B). In addition, we carried out RIP and RNA pull-down assays to explore the interaction between SNHG3 and miR-339-5p. It was turned out that miR-339-5p and SNHG3 could be precipitated by Ago2 antibody and their expression was higher than that of IgG antibody (Figure 3C), and the RNA pull-down assay revealed that SNHG3 could pull down miR-339-5p (Figure 3D). Afterwards, we quantified miR-339-5p in SKOV3 and HeyA8 cells transfected with pcDNA3.1-SNHG3, si-SNHG3, and si-NC, respectively. The qRT-PCR assay revealed that compared with SKOV3 and HeyA8 cells transfected with si-NC, those transfected with pcDNA3.1-SNHG3 showed inhibited miR-339-5p expression, and those transfected with si-SNHG3 showed up-regulated expression of it (Figure 3E). MiR-339-5p was underexpressed in OC tissues (Figure 3F) and negatively correlated with SNHG3 (Figure 3G). Moreover, we also quantified miR-339-5p in SKOV3, HeyA8, A2780, and IOSE cells, finding that the miR-339-5p expression in OC cells was significantly lower than that in IOSE cells (Figure 3H).
MiR-339-5p Regulates OC Cell Growth
The effect of miR-339-5p on the biological behavior of OC cells has not been reported before. Therefore, with the aim of studying the biological function of miR-339-5p in OC, we transfected miR-NC, miR-339-5p-mimics, and miR-339-5p-inhibitor into SKOV3 and HeyA8 cells. As shown in Figure 4A, the transfection efficiency was satisfactory. The CCK-8 assay showed that the proliferation of SKOV3 and HeyA8 cells was weakened under overexpression of miR-339-5p, while it was enhanced under knockout of miR-339-5p (Figure 4B). Additionally, the transwell assay showed that overexpressing miR-339-5p inhibited the invasion and migration of SKOV3 and HeyA8 cells, while knocking down it enhanced the abilities of cells (Figure 4C and D), and the flow cytometry analysis revealed that overexpression of miR-339-5p induced apoptosis of SKOV3 and HeyA8 cells, while knock-down of miR-339-5p caused opposite results (Figure 4E).
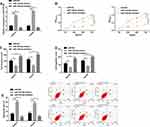
TRPC3 is the Target Gene of miR-339-5p
With the aim of exploring the potential target gene of miR-339-5p, we predicted that TRPC3 was the target gene of miR-339-5p through bioinformatics database software analysis (Figure 5A), and carried out a dual-luciferase reporter gene assay. It came out that overexpressing miR-339-5p suppressed the luciferase activity of TRPC3-WT in HeyA8 cells, while knocking down miR-339-5p enhanced it, but exerted no effect on that of TRPC3-MUT (Figure 5B). We determined the expression of TRPC3 in SKOV3 and HeyA8 cells, finding that the expression of TRPC3 was inhibited under overexpression of miR-339-5p, while it was significantly up-regulated under knock-down of miR-339-5p (Figure 5C). Subsequently, we detected the expression of TRPC3 in OC tissues by qRT-PCR. It was found that TRPC3 was overexpressed in OC tissues (Figure 5D), and negatively linked to miR-339-5p (Figure 5E), but positively correlated with SNHG3 (Figure 5F), and it was also found that TRPC3 was highly expressed in OC cells (Figure 5G).
SNHG3 Can Regulate the Target Gene of miR-339-5p, TRPC3, in OC Cells
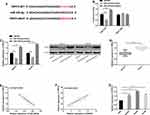
We carried out a rescue experiment to understand the role of SNHG3/miR-339-5p/TRPC3 in the growth of OC cells, and co-transfected miR-339-5p-inhibitor and pcDNA3.1-TRPC3 with si-SNHG3 into cells, respectively. Detection of the biological behavior of SKOV3 and HeyA8 cells revealed that co-transfection of si-SNHG3+miR-339-5p-inhibitor or si-SNHG3+pcDNA3.1-TRPC3 could rescue the effect of transfected si-SNHG3 on the biological behavior of SKOV3 and HeyA8 cells (Figure 6A–D). The Western blotting assay revealed that the TRPC3 protein expression could be rescued by co-transfection of si-SNHG3+miR-339-5p-inhibitor or si-SNHG3+pcDNA3.1-TRPC3 (Figure 6E).
Discussion
OC is a common malignant tumor. Despite a great progress in the diagnosis and therapy of it, the survival of patients with OC is still not optimistic.19 In our study, we verified that SNHG3, overexpressed in OC, can be adopted for diagnosis of OC, and the high expression of it indicated a poor prognosis of patients. We found that knockdown of SNHG3 could inhibit the growth of OC cells and affect the biological function of OC cells through the miR-339-5p/TRPC3 axis, which confirmed that SNHG3 is expected to be a potential marker for diagnosis, treatment and prognosis evaluation of OC.
Dysregulated lncRNA is involved in the prognosis of patients with cancers, such as gastric cancer, lung cancer, bladder cancer, and OC.20–22 Understanding the molecular mechanism of lncRNA contributes to the exploration of effective therapeutic targets for OC. For example, one study by Huang et al23 has pointed out that up-regulation of SNHG3 indicates poor prognosis of patients with colorectal cancer, and the study has also concluded that SNHG3 can act as ceRNA in colorectal cancer and promote the development of malignant tumors via the miR-182-5p/Myc pathway. One other study by Liu et al24 has pointed out that the up-regulation of SNHG3 expression in lung adenocarcinoma can accelerate the cycle, proliferation and apoptosis of lung adenocarcinoma cells, and plays a role of oncogene in tumors.
However, the role of SNHG3 in OC and its related mechanisms are still unclear. In our study, we acquired that the SNHG3 expression in tissues of patients with OC was greatly higher than that in corresponding tumor-adjacent tissues, and SNHG3 was consistently highly expressed in the serum of patients with OC, with AUC for diagnosis of OC of 0.863. In addition, we also found that high expression of SNHG3 was linked to lymph node metastasis and high histological grade of the patient, and indicated a poor prognosis of the patient, which suggests that SNHG3 participates in the development and progression of OC. Afterwards, we conducted in vitro and in vivo experiments. The in vitro experiment showed that inhibition of SNHG3 suppressed the cell viability and malignant phenotypes of the cells and accelerated apoptosis of them, and the in vivo experiment showed that inhibition of SNHG3 slowed down the growth ability of OC cells. These results imply that SNHG3 is crucial in controlling the development and progression of OC and is expected to become a potential diagnostic and therapeutic marker for OC. However, we are still not clear about its specific mechanism.
Previous studies have verified that lncRNA extensively participates in the network regulated by ceRNA and it can act as ceRNA and further play a role in miRNA sponge to regulate the transcription level of miRNA.25,26 In laryngeal carcinoma tissues and cells, SNHG3 is also significantly up-regulated, the loss of its function will reduce cell vitality, migration and invasion, and it can regulate the malignant behavior of laryngeal carcinoma cells via miR-384/WEE1 axis.27 In osteosarcoma, the up-regulation of SNHG3 indicates that the survival of patients with osteosarcoma is very poor and the up-regulation of it can cause miR-196a-5p spongization to promote malignant growth of osteosarcoma cells.28 In our study, we analyzed whether SNHG3 can interact with miRNA to act as ceRNA, and found that miR-339-5p can bind to the 3ʹUTR of SNHG3. We carried out dual-luciferase reporter gene, RIP and RNA pull-down assays, finding the interaction between SNHG3 and miR-339-5p. One previous study by Zhang et al29 has identified miRNA common in OC cells through microarray data, confirming that miR-339-5p is under-expressed in OC cells and can suppress tumor.29 Consistently, we detected a significant down-regulation of miR-339-5p in OC tissues and cells and found a negative relation between miR-339-5p and SNHG3. Subsequently, we further verified the biological behavior of miR-339-5p in OC cells, finding that knockout of miR-339-5p could suppress the malignant phenotypes of OC cells or promote cell growth. These results indicate that SNHG3 may be related to miR-339-5p and participate in the development of OC.
There are a large number of studies indicating that miR can regulate tumor growth by binding to downstream target genes.30 We further studied the relevant mechanisms of miR-339-5p in OC, and confirmed it through database. It was verified that TRPC3 was the target gene of miR-339-5p. It is consistent with the finding that there is a relation between TRPC3 and miR-339-5p in oral leukoplakia based on the analysis of miRNA expression profiles.31 TRPC3 is one of the members of transient receptor potential ion channel superfamily, and participates in the progression of cancers.32 For example, in triple-negative breast cancer, TRPC3 can regulate cell proliferation and apoptosis resistance via RASA4/MAPK signal cascade reaction.33 In melanoma, SNHG5 and TRPC3 are highly expressed, in which SNHG5 can make miR-26a-5p sponge target to TRPC3 and further promote melanoma cell growth. Inhibition of RPC3 or knockout of SNHG5 can suppress the development of tumors.34 In this study, we found that the expression of TRPC3 was inhibited after up-regulation of miR-339-5p, and we also found up-regulation of TRPC3 expression in OC tissues and cells, negative correlation of the TRPC3 expression with miR-339-5p, and positive correlation of it with SNHG5 in OC tissues. Similarly, one study by Yang et al35 has confirmed that the expression of TRPC3 in human OC tissues and cells significantly increased, and inhibition of TRPC3 in cells can suppress cell proliferation and tumor growth. We further found through rescue experiments that co-transfection of si-SNHG3+miR-339-5p-inhibitor or si-SNHG3+pcDNA3.1-TRPC3 can rescue the effect of knock-down of transfected SNHG3 on the biological behavior of OC cells and the expression of TRPC3 protein. It implies that SNHG3 can sponge miR-339-5p and participate in the biological behavior of OC cells by regulating TRPC3. Studies have shown that TRPC channel and its splice variants are important in the growth of OC cells.36 We need to further analyze the influence of TRPC3 on the activity of OC cell channel in future studies. In addition, our research has not carried out subcellular localization of SNGH3, and has not explored other interacting molecules of SNGH3, so it is still necessary to further explore the biological function and potential mechanism of SNGH3 in the future.
To sum up, SNHG3 can be adopted as a marker for diagnosis and prognosis evaluation of OC, and it can participate in the development of OC by enabling the miR-339-5p sponge to affect the TRPC3 expression. Therefore, targeting the SNHG3/miR-339-5p/TRPC3 axis is expected to be a promising therapeutic strategy for OC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lomnytska M, Pinto R, Becker S, et al. Platelet protein biomarker panel for ovarian cancer diagnosis. Biomark Res. 2018;6(1):29344361. doi:10.1186/s40364-018-0118-y
2. Montagnana M, Benati M, Danese E. Circulating biomarkers in epithelial ovarian cancer diagnosis: from present to future perspective. Ann Transl Med. 2017;5(13):276. doi:10.21037/atm.2017.05.13
3. Elias KM, Fendler W, Stawiski K, et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. Elife. 2017;6:
4. Wang H, Fu Z, Dai C, et al. LncRNAs expression profiling in normal ovary, benign ovarian cyst and malignant epithelial ovarian cancer. Sci Rep. 2016;6:38983. doi:10.1038/srep38983
5. Dong P, Xiong Y, Watari H, et al. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J Exp Clin Cancer Res. 2016;35(1):132. doi:10.1186/s13046-016-0415-y
6. Chiu HS, Somvanshi S, Patel E, et al. Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 2018;23(1):297–312 e212.
7. Chen X, Dai M, Zhu H, et al. Evaluation on the diagnostic and prognostic values of long non-coding RNA BLACAT1 in common types of human cancer. Mol Cancer. 2017;16(1):160. doi:10.1186/s12943-017-0728-2
8. Wang J, Su Z, Lu S, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. doi:10.1016/j.cca.2018.07.004
9. Zhu D, Huang X, Liang F, Zhao L. LncRNA miR503HG interacts with miR-31-5p through multiple ways to regulate cancer cell invasion and migration in ovarian cancer. J Ovarian Res. 2020;13(1):3. doi:10.1186/s13048-019-0599-9
10. Wang Y, Zhu P, Luo J, et al. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019;38(17):e101110. doi:10.15252/embj.2018101110
11. Xuan Y, Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10(10):694. doi:10.1038/s41419-019-1940-3
12. Ma Q, Qi X, Lin X, Li L, Chen L, Hu W. LncRNA SNHG3 promotes cell proliferation and invasion through the miR-384/hepatoma-derived growth factor axis in breast cancer. Hum Cell. 2020;33(1):232–242. doi:10.1007/s13577-019-00287-9
13. Zhang T, Cao C, Wu D, Liu L. SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma. Tumour Biol. 2016;37(2):2379–2385. doi:10.1007/s13277-015-4052-4
14. Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150(2):343–354. doi:10.1016/j.ygyno.2018.06.013
15. Zheng S, Jiang F, Ge D, et al. LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed Pharmacother. 2019;112:108695. doi:10.1016/j.biopha.2019.108695
16. Matz M, Coleman MP, Carreira H, et al. Worldwide comparison of ovarian cancer survival: histological group and stage at diagnosis (CONCORD-2). Gynecol Oncol. 2017;144(2):396–404.
17. Jia G, Tang Y, Deng G, et al. miR-590-5p promotes liver cancer growth and chemotherapy resistance through directly targeting FOXO1. Am J Transl Res. 2019;11(4):2181–2193.
18. Lian H, Xie P, Yin N, et al. Linc00460 promotes osteosarcoma progression via miR-1224-5p/FADS1 axis. Life Sci. 2019;233:116757. doi:10.1016/j.lfs.2019.116757
19. Qin YY, Wu YY, Xian XY, et al. Single and combined use of red cell distribution width, mean platelet volume, and cancer antigen 125 for differential diagnosis of ovarian cancer and benign ovarian tumors. J Ovarian Res. 2018;11(1):10. doi:10.1186/s13048-018-0382-3
20. Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33(4):706–720 e709. doi:10.1016/j.ccell.2018.03.006
21. Chen JF, Wu P, Xia R, et al. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17(1):6.
22. Luo H, Xu C, Le W, Ge B, Wang T. lncRNA CASC11 promotes cancer cell proliferation in bladder cancer through miRNA-150. J Cell Biochem. 2019;120(8):13487–13493. doi:10.1002/jcb.28622
23. Huang W, Tian Y, Dong S, et al. The long non-coding RNA SNHG3 functions as a competing endogenous RNA to promote malignant development of colorectal cancer. Oncol Rep. 2017;38(3):1402–1410. doi:10.3892/or.2017.5837
24. Liu L, Ni J, He X. Upregulation of the long noncoding RNA SNHG3 promotes lung adenocarcinoma proliferation. Dis Markers. 2018;2018:5736716. doi:10.1155/2018/5736716
25. Chen DL, Lu YX, Zhang JX, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7(19):4836–4849. doi:10.7150/thno.20942
26. Russo F, Fiscon G, Conte F, Rizzo M, Paci P, Pellegrini M. Interplay between long noncoding RNAs and microRNAs in cancer. Methods Mol Biol. 2018;1819:75–92.
27. Wang L, Su K, Wu H, Li J, Song D. LncRNA SNHG3 regulates laryngeal carcinoma proliferation and migration by modulating the miR-384/WEE1 axis. Life Sci. 2019;232:116597. doi:10.1016/j.lfs.2019.116597
28. Chen J, Wu Z, Zhang Y. LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma. Int J Immunopathol Pharmacol. 2019;33:2058738418820743. doi:10.1177/2058738418820743
29. Zhang S, Zhang X, Fu X, Li W, Xing S, Yang Y. Identification of common differentially-expressed miRNAs in ovarian cancer cells and their exosomes compared with normal ovarian surface epithelial cell cells. Oncol Lett. 2018;16(2):2391–2401.
30. Zhou RS, Zhang EX, Sun QF, et al. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19(1):779. doi:10.1186/s12885-019-5983-8
31. Maimaiti A, Abudoukeremu K, Tie L, Pan Y, Li X. MicroRNA expression profiling and functional annotation analysis of their targets associated with the malignant transformation of oral leukoplakia. Gene. 2015;558(2):271–277. doi:10.1016/j.gene.2015.01.004
32. Chen J, Luan Y, Yu R, Zhang Z, Zhang J, Wang W. Transient receptor potential (TRP) channels, promising potential diagnostic and therapeutic tools for cancer. Biosci Trends. 2014;8(1):1–10. doi:10.5582/bst.8.1
33. Wang Y, Qi YX, Qi Z, Tsang SY. TRPC3 regulates the proliferation and apoptosis resistance of triple negative breast cancer cells through the TRPC3/RASA4/MAPK pathway. Cancers (Basel). 2019;11(4).
34. Gao J, Zeng K, Liu Y, Gao L, Liu L. LncRNA SNHG5 promotes growth and invasion in melanoma by regulating the miR-26a-5p/TRPC3 pathway. Onco Targets Ther. 2019;12:169–179. doi:10.2147/OTT.S184078
35. Yang SL, Cao Q, Zhou KC, Feng YJ, Wang YZ. Transient receptor potential channel C3 contributes to the progression of human ovarian cancer. Oncogene. 2009;28(10):1320–1328. doi:10.1038/onc.2008.475
36. Zeng B, Yuan C, Yang X, Atkin SL, Xu SZ. TRPC channels and their splice variants are essential for promoting human ovarian cancer cell proliferation and tumorigenesis. Curr Cancer Drug Targets. 2013;13(1):103–116. doi:10.2174/156800913804486629
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.