Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA ZNF667-AS1 Promotes ABLIM1 Expression by Adsorbing microRNA-1290 to Suppress Nasopharyngeal Carcinoma Cell Progression
Authors Chen X, Huang Y, Shi D, Nie C, Luo Y, Guo L, Zou Y, Xie C
Received 10 January 2020
Accepted for publication 15 April 2020
Published 20 May 2020 Volume 2020:13 Pages 4397—4409
DOI https://doi.org/10.2147/OTT.S245554
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Xi Chen,1,2 Yaping Huang,1 Dianyu Shi,2 Chuan Nie,3 Yiping Luo,4 Liangfen Guo,1 Yu Zou,1 Chun Xie5
1Department of Otorhinolaryngology, Guangdong Women and Children Hospital, Guang Zhou, Guangdong, 511400, People’s Republic of China; 2Department of Otorhinolaryngology, People’s Hospital of Longhua,Guangdong,People’s Republic of China; 3Department of Neonatology, Guangdong Women and Children Hospital, Guang Zhou 511400, Guangdong, People’s Republic of China; 4Department of Internal Medicine, Guangdong Women and Children Hospital, Guang Zhou 511400, Guangdong, People’s Republic of China; 5Department of Stomatology, People’s Hospital of Longhua, Shenzhen 518109, Guangdong, People’s Republic of China
Correspondence: Yu Zou; Chun Xie Email [email protected]; [email protected]
Background: Recently, long non-coding RNAs (lncRNAs) have been elucidated to play essential roles in cancers, and the recognition of lncRNA expression patterns in nasopharyngeal carcinoma (NPC) may be helpful for indicating novel mechanisms underlying NPC carcinogenesis. Herein, we conducted this study to probe into the function of lncRNA ZNF667-AS1 in NPC progression with the involvement of microRNA-1290 (miR-1290) and actin-binding LIM protein 1 (ABLIM1).
Materials and Methods: In silico analysis screened differentially expressed genes and miRNAs in NPC and predicted potential mechanisms. ZNF667-AS1 expression was detected in NPC tissues and cells. The gain-and-loss function assays were performed to explore the effects of lncRNA ZNF667-AS1 and miR-1290 in NPC cell biological behaviors. In vivo experiments were further conducted to confirm the in vitro results.
Results: In silico analysis predicted that ZNF667-AS1 was diminished in NPC, which may downregulate ABLIM1 through sponging miR-1290. ZNF667-AS1 was poorly expressed in NPC tissues and cells, and overexpression of ZNF667-AS1 inhibited growth of NPC cells. ZNF667-AS1 competitively bound with miR-1290, thereby upregulating ABLIM1. miR-1290 resulted in the promotion of NPC cell progression by suppressing ABLIM1. Overexpression of ZNF667-AS1 or suppression of miR-1290 inhibited tumorigenicity of NPC cells in vivo.
Conclusion: This study highlights that lncRNA ZNF667-AS1 promotes ABLIM1 expression by sponging miR-1290 to suppress NPC cell progression.
Keywords: nasopharyngeal carcinoma, lncRNA ZNF667-AS1, ABLIM1, microRNA-1290, competing endogenous RNA
Introduction
Nasopharyngeal carcinoma (NPC) is defined as a type of malignant tumor occurring in head and neck, and it develops in the nasopharynx’s epithelial lining.1 NPC is common in south-eastern Asia, Arctic region and China, with the incidence of 30 per 100,000.2 The risk factors of NPC consist of three main reasons: viral infection, genetic factors as well as environmental and dietary factors.3 Except for active anticancer agents and intensity-modulated radiation therapy, cancer stem cells together with gene therapy are considered as novel concepts and promising regimens for the treatment of NPC, while these technologies have yet been put into clinical use.4 In view of the lack of reliable data, the pathogenesis and clinical use of NPC biomarkers remain to be uncovered.5 As such, it is crucial to recognize novel biomarkers and to develop novel effective treatments of NPC through exploring the molecular mechanisms in NPC.
Long non-coding RNAs (lncRNAs) have been elucidated to play a part in different cancers, and evidence has concentrated on the functions of lncRNAs in malignant behaviors of various cancers.6 Emerging evidence has indicated that some lncRNAs may influence NPC growth, such as lncRNA PCAT7, lncRNA ROR, and lncRNA ANRIL.7–9 LncRNA ZNF667-AS1, once named MORT, is expressed in all human cells, while its deficiency is observed in human breast epithelial cells.10 A study has shown that a reduction of ZNF667-AS1 together with ZNF667 was found in esophageal cancer cells and esophageal squamous cell carcinoma (ESCC) tissues.11 In general, lncRNAs are implicated in malignant phenotypes of cancer cells via altering expression of targeted genes through varying mechanisms, the most important one is sponging microRNAs (miRNAs).8 A recent article has revealed dysregulation of some miRNAs in NPC, and have also found clinically prognostic miRNA signatures.12 miR-1290 expression is overexpressed, which is related to disease progression in ESCC patients.13 Interestingly, miRNAs play vital roles in the post-transcriptional gene modulation through the 3ʹuntranslated region (UTR) of target mRNAs.14 Actin-binding LIM protein 1 (ABLIM1) has been described to be positioned in a genomic area frequently depleted in human tumors, which is implicated in axon guidance.15 Evidence has shown that ABLIM1 is markedly lower in adrenocortical carcinomas in comparison to adrenocortical adenomas.16 Nevertheless, the impacts of the ZNF667-AS1/miR-1290/ABLIM1 axis on the development of NPC remain to be addressed. As a consequence, we performed this present study for validation.
Materials and Methods
Ethics Statement
This experiment was implemented with the approval of the ethics committee of People’s Hospital of Longhua. All the participants offered the written informed consent. All protocols were performed following the ethical principles for medical research regarding human subjects of the Declaration of Helsinki. The current work was carried out in strict adherence to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was also approved by the Institutional Animal Care and Use Committee of the People’s Hospital of Longhua. Significant efforts were made for eliminating the pain suffered by the animals.
In silico Analysis
The Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) was searched to find NPC-related gene (GSE12452) and miRNA (GSE70970) microarray datasets. The R language limma package (http://master.bioconductor.org/packages/release/bioc/html/limma.html) was searched to analyze the expression matrix of the microarray datasets. The screening conditions for differentially expressed genes (DEGs) were adj.p.Val <0.05 and |LogFoldChange | >1. R language pheatmap package (https://cran.r-project.org/web/packages/pheatmap/index.html) was utilized to plot a heatmap for DEGs. RNA22 (https://cm.jefferson.edu/rna22/) website, a database could predict the binding sites between RNAs, was used to predict the relationship between the differentially expressed lncRNA and miRNA. In addition, miRDB (http://www.mirdb.org/), mirDIP (http://ophid.utoronto.ca/mirDIP/), DIANA, and Targetscan (http://www.targetscan.org/vert_71/) could forecast the targeting genes for miRNA. The jvenn (http://jvenn.toulouse.inra.fr/app/example.html) could compare and analyze different element datasets and draw Venn diagrams.
Study Subjects
From November 2017 to November 2018, nasopharyngeal biopsies and adjacent tissues of 36 NPC patients (14 males and 22 females, aged 28–71 years) were collected from People’s Hospital of Longhua. All patients were diagnosed by pathological examination and none received any chemotherapy or radiotherapy. None of the patients had a history of other malignancies.
Cell Treatment and Grouping
Human nasopharyngeal epithelial cells NP69 (American Type Culture Collection (ATCC), Manassas, VA, USA) and four NPC cell lines c666-1 (ATCC), CNE-1, CNE-2 and HNE1 (Zhongqiao Xinzhou Biotechnology Co., Ltd., Shanghai, China) were cultured in Roswell Park Memorial Institute (RPMI)-1640 supplemented with 10% fetal bovine serum (FBS, Gibco Company, Grand Island, NY, USA), penicillin (100 U/mL) and streptomycin (100 mg/mL) at 37ºC with 5% CO2. Based on the cell growth situation, the cells were subcultured when reaching 85% or higher confluence.
Cells were seeded in 6-well plates at 24 h pre-transfection with 30–50% cell density. Cells were transfected in the light of instructions of the lipofectamine 2000 reagent (11668–019, Invitrogen, New York, CA, USA). After cells in each group were incubated with 5% CO2 for 6–8 h, the complete medium was renewed. After a 48-h culture, further experiments were performed. The cells were introduced with overexpressed (oe)-ZNF667-AS1, short hairpin RNA (sh)-ZNF667-AS1, miR-1290 mimic, miR-1290 inhibitor or their negative controls (NCs). Overexpression plasmids, shRNA, inhibitors and mimics were purchased from GenePharma (Shanghai, China). Sequence is shown in Table 1.
 |
Table 1 Sequence for Cell Transfection |
Dual-Luciferase Reporter Gene Assay
An ABLIM1-3ʹuntranslated region (3ʹUTR) fragment with a miR-1290 binding site and a ZNF667-AS1 cDNA fragment were inserted into the pGL3 plasmid. With a point mutation method, ABLIM1-3ʹUTR-mutant type (MUT) and ZNF667-AS1-MUT fragments were also inserted into the pGL3 plasmid. Using liposome transfection method, pGL3-ZNF667-AS1, pGL3-lncRNA ZNF667-AS1-MUT, pGL3-ABLIM1-3ʹUTR, pGL3-ABLIM1-3ʹUTR-MUT recombinant vector and Renilla internal reference plasmid were co-transfected with miR-1290 mimic or NC-mimic into HEK293T cells. At 48 hours post-transfection, cells were harvested and lysed, and the luciferase activity was determined by a luciferase detection kit (K801-200, BioVision, Exton, PA, USA) and a dual-luciferase reporter gene analysis system (Promega, Madison, WI, USA).
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
The culture plate was incubated with EdU solution, fixed for 30 min with paraformaldehyde (40 g/L), and incubated for 8 min with glycine solution. Next, the culture plate was rinsed with phosphate buffered saline (PBS) containing 0.5% TritonX-100, stained with Apollo® staining reaction solution under conditions void of light. Finally, the cells were stained with Hoechst 3334 reaction solution in darkness and observed under a fluorescent microscope. EdU-stained cells and Hoechst 33342-stained cells were counted under three fields. Cell proliferation rate was calculated as the number of EdU-stained cells/Hoechst 33342-stained cells × 100%.
Scratch Test
The cells were seeded into 6-well plates at 48 h post-transfection with 5 × 105 cells in each well. When reaching about 90% cell confluence, the cells were scratched with a sterile pipette tip cross the middle axis of the well. After the removal of the floating cells, the remaining cells were further cultured for 0.5–1 h in serum-free medium for recovery. The images were captured at 0 h and 24 h, and the migration distance was gauged by the Image-Pro Plus Analyses software (Media Cybernetics, Silver Spring, MA, USA).
Transwell Assay
Matrigel (356234, BD Biosciences, Franklin Lakes, NJ, USA) was dissolved 48 h post-transfection, diluted at a ratio of 1:3 with serum-free medium and added to the apical chamber at 50 μL/well. The cells were detached and dispersed into cell suspension. Afterwards, the cell suspension (1 × 105 cells/mL) was seeded into the apical chamber. The basolateral chamber was incubated with medium containing 10% FBS at 37°C for 24 h. Transwell chamber was fastened with 5% glutaraldehyde, and stained with 0.1% crystal violet staining solution for 30 min and observed under a microscope. The cell number that passed through Matrigel in every group was utilized as an index to evaluate their invasion ability.
Flow Cytometry
Forty-eight hours post-transfection, cells were trypsinized with 0.25% trypsin to adjust the concentration for 1 × 106 cells/mL. The cells (1 mL) were centrifuged, with the supernatant removed. After that, the cells were fixed with 70% ethanol at 4°C overnight, and 100 μL cell suspension was stained with 50 μg RNAase-containing propidium iodide (PI) staining solution (40710ES03, Qianchen Biotechnology, Shanghai, China). After 30 min in the dark, the cells were filtered with a 100-mesh nylon. A flow cytometer (BD, FL, NJ, USA) was adopted to detect the cell cycle by recording the red fluorescence at 488 nm.
After 48-h incubation, the cells were centrifuged to resuspend in the binding buffer (200 μL). The cells were mixed with 10 μL Annexin V-fluorescein isothiocyanate (ab14085, Abcam, Inc, MA, USA) and 5 μL PI, and then added with 300 μL binding buffer. Finally, the cell apoptosis was determined with a 488 nm excitation wavelength by a flow cytometer.
Fluorescence in situ Hybridization (FISH)
FISH technique was carried out to recognize the subcellular localization of lncRNA ZNF667-AS1. Based upon the RiboTM lncRNA FISH probe Mix (Red, Ribobio, Guangzhou, China) instructions, a coverslip was positioned in a 6-well culture plate. NPC cells were seeded and cultured to achieve a cell confluence of about 80%. Next, the cells were fixed with 4% paraformaldehyde. After treatment with proteinase K (2 μg/mL), glycine, and acetylcarnitine, the cells were successively incubated with 250 μL prehybridization solution at 42°C for 1 h and 250 μL hybridization solution containing probe (300 ng/mL) overnight. Subsequently, cells were stained with 6-diamidino-2-phenylindole for 5 min. After washing, cell coverslips were sealed with anti-fluorescence quenching agent and photographed under a fluorescence microscope (Olympus, Tokyo, Japan) by selecting five different horizons.
RNA Immunoprecipitation (RIP) Assay
LncRNA ZNF667-AS1 bound to AGO2 protein (rabbit anti-AGO2, ab32381, 1:50) and IgG (ab109489, 1:100, both Abcam) was determined with a RIP kit (Millipore, Billerica, MA, USA). With the supernatant removed, NPC cells were lysed with radio-immunoprecipitation assay lysis buffer (P0013B, Beyotime, Shanghai, China), and centrifuged. Briefly, the magnetic beads (50 μL) were resuspended in RIP wash buffer (100 μL), and 5 μg antibody was supplemented based on the grouping for binding. Next, the magnetic beads-antibody complex after washing were resuspended in 900 μL RIP wash buffer, and the cell extract (100 μL) was supplemented for incubation. After that, the magnetic beads-protein complex was amassed, and the sample and input were trypsinized with proteinase K for RNA extraction for the subsequent reverse transcription quantitative polymerase chain reaction (RT-qPCR).
RNA Pull-Down Assay
NPC cells were transduced with 20 nM biotinylated wild type (WT)-bio-miR-1290 and MUT-bio-miR-1290. Forty-eight hours later, the cells were harvested and washed. According to the instructions of the kit, 20 mL streptavidin-coated magnetic beads (Dynabeads M-280, 11206D, Life Technology, Carlsbad, CA, USA) were activated. The magnetic beads were sealed with 10 mg/mL RNase-free bovine serum albumin and yeast tRNA (R8508, Sigma-Aldrich Chemical Company, St Louis, MO, USA) at 4°C for 30 min. The cells were then incubated for 10 min with a specific lysis buffer (Ambion, Austin, TX, USA). Next, the lysates experienced an incubation with M-280 streptavidin magnetic beads (Sigma, St Louis, MO, USA) for 2 h for RNA extraction. The bound RNA was purified by Trizol and the lncRNA ZNF667-AS1 enrichment was determined by RT-qPCR.
Western Blot Analysis
The total protein was ice-bathed with lysis buffer and proteinase inhibitor (1111111, Beijing Jiamei Niunuo Biotechnology, Beijing, China) for 30 min and centrifuged for 10 min. The protein (50 μg) experienced electrophoresis, and transferred onto a polyvinylidene difluoride membrane and then sealed with 5% skimmed milk. Subsequently, the membrane was probed with diluted primary antibodies: ABLIM1 (1:1000, ab222824), proliferating cell nuclear antigen (PCNA, 1:1000, ab18197), Bcl-2 (1:1000, ab32124), Bax (1:2000, ab32503), N-cadherin (1:1000, ab18203), matrix metalloprotease (MMP)-9 (1:1000, ab138898), glyceraldehyde phosphate dehydrogenase (GAPDH, 1:2000, ab9485) (Abcam) and then with goat anti-rat horseradish peroxidase-conjectured secondary antibody against IgG (HA1003, Yanhui Bio, Shanghai, China). Lastly, the membrane was colored by enhanced chemiluminescence reagent (ECL808-25, Biomiga, San Diego, CA, USA), and the relative expression of the protein was determined.
RT-qPCR
Total RNA was acquired using the miRNeasy Mini Kit (217004, QIAGEN, Hilden, Germany). The primer was both designed and synthetized by Takara Company (Dalian, Liaoning, China) (Table 2). Next, RNA was reversely transcribed into cDNA with the PrimeScript RT kit (RR036A, Takara, Kyoto, Japan). RT-qPCR was implemented based upon a SYBR® Premix Ex TaqTM II Kit (RR820A, Takara) according to the manufacturer’s requirements using the ABI 7500 PCR instrument (ABI, Oyster Bay, N.Y., USA). U6 and GAPDH were regarded as loading controls. On the basis of the threshold cycle (Ct), the expression of target genes relative to a loading control was calculated using the formula 2−ΔCT, where ΔCT = (Ct target gene – Ct loading control) and ∆∆CT = (∆Ct the model group - ∆Ct the normal group). The formula 2−∆∆CT was calculated to demonstrate the relative expression of each gene compared to the loading control.
 |
Table 2 Primer Sequence |
Tumor Xenografts in Nude Mice
BALB/c nude mice (3–4 w, 14 ± 2 g) were given free access to food and water during the experiments. The temperature was maintained at about 22-27°C and humidity between 45% and 50% throughout the study. The tumorigenic ability of the six groups was detected: oe-NC, oe-ZNF667-AS1, inhibitor NC, miR-1290 inhibitor, oe-ZNF667-AS1 + sh-NC and oe-ZNF667-AS1 + sh-ABLIM1A (n = 3). A stably transfected cell line was constructed, and the cell concentration was altered to 1 × 107 cells/mL. The cell suspension (20 μL) was injected subcutaneously in nude mice, and the tumor growth was observed. The tumor volume was recorded every 7 d to plot the growth curve: (a*b2)/2 (a, longest diameter; b, shortest diameter). Mice were euthanized 35 days later with carbon dioxide asphyxia, and the tumors were weighed.
Statistical Analysis
SPSS 21.0 software (IBM Corp., Armonk, NY, USA) was applied for data analysis. The results were presented in the form of mean ± standard deviation. The paired t test was employed for analysis of comparisons between tumor tissues and adjacent tissues, while the unpaired t test for other pairwise comparisons. One-way analysis of variance (ANOVA) was adopted for comparisons among multi-groups, and Tukey’s post hoc test for pairwise comparisons after ANOVA. p < 0.05 indicated significant difference.
Results
ZNF667-AS1 is Reduced in NPC Tissues and Cells
Firstly, differential analysis was performed on the NPC-related GSE12452 microarray dataset, and 10 differently expressed lncRNAs were screened out, and a heatmap of those lncRNAs is displayed in Figure 1A. We have noticed that ZNF667-AS1 was downregulated in NPC tissues, and some studies have shown that ZNF667-AS1 is poorly expressed in cervical cancer17 and esophageal squamous cell carcinoma,18 but the significance in NPC is not clear. Therefore, we studied the potential impact of ZNF667-AS1 on NPC.
Identical to the database prediction results, RT-qPCR results indicated that ZNF667-AS1 was poorly expressed in NPC tissues (p < 0.05; Figure 1B). In addition, ZNF667-AS1 in NPC cells were also determined, which suggested that versus nasopharyngeal epithelial cell NP69, the ZNF667-AS1 expression in c666-1, HNE1, CNE-1 and CNE-2 cells was reduced, and ZNF667-AS1 was the lowest in CNE-1 cells (p < 0.05; Figure 1C), which was selected for subsequent experiments.
Overexpression of ZNF667-AS1 Inhibits Progression of NPC Cells
The next step was to determine the efficiency of ZNF667-AS1 overexpression or knockdown transfection, and the results illustrated that ZNF667-AS expression increased in NPC cells with oe-ZNF667-AS, while reduced in cells upon sh-ZNF667-AS1-1, sh-ZNF667-AS1-2, and sh-ZNF667-AS1-3 treatment. Furthermore, ZNF667-AS1 was the lowest in the sh-ZNF667-AS1-1 group, so it was chosen for subsequent experiments (Figure 2A).
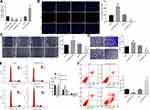
For verifying the role of ZNF667-AS1 in biological functions of CNE-1 cells, we identified that overexpressed ZNF667-AS1 diminished the proliferation rate, the migration distance at 48 h, and the number of invading cells, arrested cells at G0/G1 phase, as well as elevated apoptosis rate in CNE-1 cells. On the contrary, silencing of ZNF667-AS1 presented an opposite tendency (Figure 2B–F). The above results show that overexpression of ZNF667-AS1 inhibits the proliferation, migration and invasion abilities of NPC cells and affects the cell cycle to promote their apoptosis.
ZNF667-AS1 Competitively Interacts with miR-1290
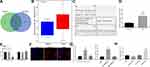
A differential analysis was performed on the NPC miRNA GSE70970 microarray dataset. Twelve miRNAs were found to be overexpressed in NPC. In addition, RNA22 was utilized to predict the miRNAs that might bind to ZNF667-AS1, and the miRNA prediction result was compared with the up-regulated miRNAs in the GSE70970 microarray dataset. Two miRNAs, namely hsa-miR-1290 and hsa-miR-1275, were identified in the intersection (Figure 3A). Evidence has reported the role of miR-1275 in NPC,19 while the function of miR-1290 on NPC development was rarely investigated. Meanwhile, miR-1290 in GSE70970 was highly expressed in NPC tissues (Figure 3B). RNA22 predicted that there was a putative binding site between ZNF667-AS1 and miR-1290 (Figure 3C). It is speculated that the miR-1290 in NPC may be regulated by ZNF667-AS1. Consistently, miR-1290 was detected to be highly expressed in NPC tissues from RT-qPCR results (Figure 3D). As shown in Figure 3E, we found that the miR-1290 mimic abated the luciferase activity of the ZNF667-AS1-WT, while it had no impact on the luciferase activity of the ZNF667-AS1-MUT, indicating that ZNF667-AS1 bound to miR-1290.
Subsequently, the experimental results of FISH technique are shown in Figure 3F: the blue part corresponded to the nucleus and the red part, ZNF667-AS1, indicating that ZNF667-AS1 was largely presented in the cytoplasm. We, therefore, conjectured that ZNF667-AS1 could modulate NPC progression by modulating miR-1290. As shown in RNA-pull down assay (Figure 3G), the ZNF667-AS1 expression increased in the WT-miR-1290 group, suggesting that there was a direct binding relationship between miR-1290 and ZNF667-AS1. Similarly, compared with MUT-ZNF667-AS1 and Bio-NC groups, the enrichment of miR-1290 detected in the WT-ZNF667-AS1 group increased significantly, further indicating that miR-1290 and ZNF667-AS1 could bind directly. Furthermore, RIP assay indicated that in CNE-1 cells, AGO2 antibodies could precipitate ZNF667-AS1. Moreover, we found that the expression level of ZNF667-AS1 precipitated by AGO2 protein in CNE-1 cells transfected with miR-1290 inhibitor was significantly decreased, and improved in cells transfected with miR-1290 mimic, implying that ZNF667-AS1 could form a complex with AGO2 and be competitively bound to miR-1290 (Figure 3H).
miR-1290 Targets and Negatively Modulates ABLIM1 Expression
The target genes of miR-1290 were forecasted in RNA22, mirDIP, miRDB, DIANA, and Targetscan, and the predicted results were compared with genes with low expression of NPC in GSE12452 microarray dataset. There were three intersecting genes: OSBPL6, ABLIM1, CHL1 (Figure 4A). The role of OSBPL6 has been reported in Alzheimer’s disease,20 but rarely examined in cancers. Besides, CHL1 deletion has been revealed in NPC with the involvement of the PI3K/AKT signaling.21 Meanwhile, ABLIM1 was reported to be a tumor suppressor, but its effect on NPC was not clear. The expression of ABLIM1 in NPC tissues in GSE12452 microarray dataset was downregulated (Figure 4B). The binding site between miR-190 and ABLIM1 was predicted using RNA22 (Figure 4C). In summary, we speculate that ZNF667-AS1 may regulate ABLIM1 through miR-1290 in NPC.
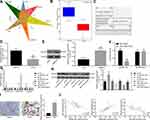 |
Figure 4 ABLIM1 is a direct target of miR-1290. (A) The target genes of miR-1290 were predicted in RNA22, mirDIP (http://ophid.utoronto.ca/mirDIP/), miRDB (http://www.mirdb.org/), DIANA (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index), and TargetScan (http://www.targetscan.org/vert71/), and the predicted results of target genes were compared with genes with low expression of NPC in GSE12452 microarray dataset. There were three intersecting genes: OSBPL6, ABLIM1, CHL1. (B) The expression of ABLIM1 in the GSE12452 microarray dataset. (C) RNA22 predicts the binding site between miR-1290 and ABLIM1. (D) RT-qPCR for the detection of ABLIM1 mRNA expression in NPC and adjacent tissues (n = 36). (E) Western blot analysis for the detection of ABLIM1 protein expression in NPC and adjacent tissues (n = 36), * p < 0.05 vs adjacent tissues. (F) Dual-luciferase reporter gene assay to verify the binding relationship between miR-1290 and ABLIM1. (G) After overexpression or silencing of ZNF667-AS1 and miR-1290 in CNE-1 cells, RT-qPCR was used for the detection of ZNF667-AS1, miR-1290 and ABLIM1 expression. (H) After overexpression or silencing of ZNF667-AS1 and miR-1290 in CNE-1 cells, Western blot analysis was used for the detection of ABLIM1 expression. (I) Immunohistochemical staining for the detection of ABLIM1 protein expression in NPC and adjacent tissues (n = 36). (J) Pearson’s correlation test for the interactions between ABLIM1, ZNF667-AS1 and miR-1290. *p < 0.05 vs the oe-NC group. #p < 0.05 vs sh-NC group. &p < 0.05 vs the mimic NC group. @p < 0.05 vs the inhibitor NC group. Data are expressed as mean ± standard deviation. Paired t test is used for comparison of two groups. One-way analysis of variance is used for comparison among multiple groups. Each experiment was run in triplicate. Abbreviations: ABLIM1, actin-binding LIM protein 1; miR-1290, microRNA-1290; RT-qPCR, reverse transcription quantitative polymerase chain reaction; NPC, nasopharyngeal carcinoma; ZNF667-AS1, zinc finger protein 667-antisense RNA 1; NC, negative control; oe, overexpression; shRNA, short hairpin RNA; WT, wild type; MUT, mutant; GAPDH, glyceraldehyde phosphate dehydrogenase. |
The statistical results of RT-qPCR and Western blot analysis revealed decreased expression of ABLIM1 in NPC tissues (Figure 4D and E). As shown in Figure 4F, we found that the miR-1290 mimic suppressed the luciferase activity of the ABLIM1-WT, but had no change in the ABLIM1-MUT, revealing that miR-1290 targeted ABLIM1.
Additionally, expression of ZNF667-AS1, <italic>miR-1290</italic>, and ABLIM1 was detected after ZNF667-AS1 and miR-1290 were overexpressed or silenced, respectively. The findings suggested that overexpressed ZNF667-AS1 enhanced ZNF667-AS1 and ABLIM1, while reduced miR-1290 expression; silencing of ZNF667-AS1 reduced the expression of ZNF667-AS1 and ABLIM1, while elevated miR-1290 expression. Overexpression of miR-1290 reduced ABLIM1 expression, while had no significant change in ZNF667-AS1 expression; silencing of miR-1290 decreased ABLIM1 expression, but had no significant change in ZNF667-AS1 expression (Figure 4G and H). In addition, we tested its expression in clinical samples by immunohistochemistry. The expression level of ABLIM1 in tumor tissues was significantly higher than that in adjacent tissues (Figure 4I). Therefore, to further determine the role of the ZNF667-AS1/miR-1290/ABLIM1 axis in NPC, we analyzed the correlation between miR-1290 and ABLIM1 or ZNF667-AS1 in 36 NPC patients by Pearson’s correlation test. The results showed that the expression level of ABLIM1 and ZNF667-AS1 was negatively correlated with miR-1290, and ABLIM1 was positively correlated with ZNF667-AS1 (Figure 4J).
Based on the above results, ZNF667-AS1 functions as a molecular sponge of miR-1290, thus upregulating the expression of ABLIM1.
miR-1290 Promotes Progression of NPC Cells by Suppressing ABLIM1
For confirming the roles of miR-1290 and ABLIM in biological functions of CNE-1 cells, the findings indicated that overexpressed miR-1290 increased the proliferation rate, the migration distance at 48 h, and the number of invading cells, arrested cells at S phase, as well as decreased apoptosis rate in CNE-1 cells. On the contrary, suppression of miR-1290 presented an opposite tendency. Moreover, the silencing of ABLIM reversed the suppression of the progression of NPC cells induced by suppression of miR-1290 (Figure 5A–E).
Besides, the expressions of ABLIM1, proliferation-related factor PCNA, migration-related factors N-cadherin and MMP-9, and apoptosis-related proteins Bcl-2 and Bax in each group of cells were detected. The results mirrored that overexpressed miR-1290 decreased ABLIM1 and Bax expression, while enhanced expression of PCNA, N-cadherin, MMP-9 and Bcl-2. On the contrary, suppression of miR-1290 presented an opposite trend. Moreover, silencing of ABLIM reversed the promotion of ABLIM1 and Bax expression and suppression of PCNA, N-cadherin, MMP-9 and Bcl-2 induced by suppression of miR-1290 (Figure 5F).
Overexpression of ZNF667-AS1 or Suppression of miR-1290 Inhibits Tumor Growth of NPC Cells in vivo
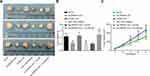
With the aim to recognize the functions of ZNF667-AS1 and miR-1290 on the tumor growth of NPC cells, we injected the stably transfected cells in nude mice and analyzed the tumor size and weight (Figure 6A–C). The obtained results revealed that overexpressed ZNF667-AS1 or suppressed miR-1290 decreased the tumor volume and weight. In addition, silencing ABLIM1 reversed the suppressive effect of ZNF667-AS1 overexpression on tumorigenicity of NPC cells in vivo.
Discussion
Despite local control has made remarkable progresses, the outcomes of NPC remain dismal due to distant failure together with late toxicity in satisfactory local control, particularly for these cancers with intracranial invasion.22 Meanwhile, it has been reported that lncRNAs may engage in the NPC mechanisms, which may also act as prognostic factors for NPC and potential targets for NPC treatment.23 As reported, miR-1290 acts as an emerging onco-miRNA which is of great importance in tumor development.24 In view of this, we launched this experiment to unmask the impacts of ZNF667-AS1/miR-1290/ABLIM1 on the inner mechanisms of NPC.
In our study, one of the findings mirrored that ZNF667-AS1 was reduced in NPC, and overexpressed ZNF667-AS1 resulted in suppressed progression of NPC cells and inhibited tumorigenicity of NPC cells in vivo. In line with our results, overexpressed ZNF667-AS1 and ZNF667 have been found to impede the viability, migration as well as invasion of ESCC cells in vitro.11 Additionally, ZNF667-AS1 has been detected to be lower in cervical cancer (CC) tissues, whose expression was inversely related to the overall survival, the proliferation and cell colony formation abilities of CC cells.25 Similarly, upregulation of ZNF667-AS1 found to suppress cell invasion and cell cycle entry in vitro and to weaken tumor metastasis in vivo.17 Another study has also indicated that ZNF667-AS1 is poorly expressed after spinal cord injury (SCI), which restricts the inflammatory response and induces SCI recovery.26
Meanwhile, our study suggested that overexpressed ZNF667-AS1 promotes ABLIM1 by interacting with miR-1290. A new model involving lncRNAs has been proposed in gene modulation, which called competing endogenous RNA.27 Based on which, lncRNAs are able to bind with miRNAs and control the expression of protein-coding gene, thereby participating in the cell behaviors. These interactions among lncRNAs, miRNAs and mRNAs produce complicated regulatory networks.28 In this current study, we determined that ZNF667-AS1 could bind to miR-1290 and directly regulate ABLIM1 expression in NPC development. Due to the inadequate evidence on this aspect, further research is needed to affirm our results.
This study also confirmed that miR-1290 targeted ABLIM1, and miR-1290 promoted the progression of NPC cells by suppressing ABLIM1. Meanwhile, suppression of miR-1290 inhibited tumorigenicity of NPC cells in vivo. A research has elucidated that the serum miR-1290 levels increased in ESCC patients, implying that serum miR-1290 could be applied as an effective diagnostic and prognostic biomarker of ESCC.29 As previously described, miR-1290 exhibited oncogenic functions, such as the enhancement of ESCC cell malignant behaviors, highlighting the significant role of miR-1290 in carcinoma progression.13 miR-1290 has also been demonstrated to induce occurrence of ESCC cells through suppressing NFIX expression, indicating that miR-1290 and NFIX might modulate biological function and affect tumor progression.30,31 Another article has manifested the pathogenic potential of miR-1290 in lung squamous cell carcinoma via the suppression of its putative target genes MAF and ITPR2.32 ABLIM1 has been revealed to be a potential bridging molecule between the signaling pathways and actin-based cytoskeleton.15 ABLIM1 expression levels have been detected to be gradually elevated with time in RANKL-induced osteoclasts in comparison to the non-induced controls.33 Besides, the knockdown of ABLIM1 has been demonstrated to promote the invasion of the metastatic human WM115 cells, whereas reciprocally, overexpression of ABLIM1 impeded the invasion of the human SKMel28 and MUM2C cells.34 Furthermore, knockdown of ABLIM1 has also been found to strengthen the forming ability of multinucleated osteoclasts, which could markedly enhance the osteoclast-marker gene expression.35 Collectively, these findings suggest that more studies are warranted to affirm the prognostic value of miR-1290 and ABLIM1 in NPC.
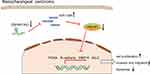
In summary, this present study indicates that overexpressed ZNF667-AS1 may promote ABLIM1 expression by binding to miR-1290, thus restricting the progression of NPC cells (Figure 7). This research delineates a new regulatory network of ZNF667-AS1/miR-1290/ABLIM1 axis in regulating the biological process and further affecting tumor growth of NPC.
Data Sharing Statement
All the data generated or analyzed in the process of this study are included in this published article.
Disclosure
All authors declare no conflicts of interest in this study.
References
1. Zhao L, Fong AHW, Liu N, Cho WCS. Molecular subtyping of nasopharyngeal carcinoma (NPC) and a microRNA-based prognostic model for distant metastasis. J Biomed Sci. 2018;25(1):16. doi:10.1186/s12929-018-0417-5
2. Zucchetto A, Taborelli M, Bosetti C, et al. Metabolic disorders and the risk of nasopharyngeal carcinoma: a case-control study in Italy. Eur J Cancer Prev. 2018;27(2):180–183. doi:10.1097/CEJ.0000000000000286
3. Roy Chattopadhyay N, Das P, Chatterjee K, Choudhuri T. Higher incidence of nasopharyngeal carcinoma in some regions in the world confers for interplay between genetic factors and external stimuli. Drug Discov Ther. 2017;11(4):170–180. doi:10.5582/ddt.2017.01030
4. Li S, Hang L, Ma Y, Wu C. Distinctive microRNA expression in early stage nasopharyngeal carcinoma patients. J Cell Mol Med. 2016;20(12):2259–2268. doi:10.1111/jcmm.12906
5. Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80. doi:10.1016/S0140-6736(19)30956-0
6. Li G, Liu Y, Liu C, et al. Genome-wide analyses of long noncoding RNA expression profiles correlated with radioresistance in nasopharyngeal carcinoma via next-generation deep sequencing. BMC Cancer. 2016;16:719. doi:10.1186/s12885-016-2755-6
7. Li L, Gu M, You B, et al. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 2016;107(9):1215–1222. doi:10.1111/cas.12989
8. Liu Y, Tao Z, Qu J, Zhou X, Zhang C. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2017;491(2):374–381. doi:10.1016/j.bbrc.2017.07.093
9. Wu JH, Tang JM, Li J, Li XW. Upregulation of SOX2-activated lncRNA ANRIL promotes nasopharyngeal carcinoma cell growth. Sci Rep. 2018;8(1):3333. doi:10.1038/s41598-018-21708-z
10. Vrba L, Garbe JC, Stampfer MR, Futscher BW. A lincRNA connected to cell mortality and epigenetically-silenced in most common human cancers. Epigenetics. 2015;10(11):1074–1083. doi:10.1080/15592294.2015.1106673
11. Dong Z, Li S, Wu X, et al. Aberrant hypermethylation-mediated downregulation of antisense lncRNA ZNF667-AS1 and its sense gene ZNF667 correlate with progression and prognosis of esophageal squamous cell carcinoma. Cell Death Dis. 2019;10(12):930. doi:10.1038/s41419-019-2171-3
12. Spence T, Bruce J, Yip KW, Liu FF. MicroRNAs in nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5(2):17. doi:10.21037/cco.2016.03.09
13. Li M, He XY, Zhang ZM, et al. MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis. World J Gastroenterol. 2015;21(11):3245–3255. doi:10.3748/wjg.v21.i11.3245
14. Bei Y, Song Y, Wang F, et al. miR-382 targeting PTEN-Akt axis promotes liver regeneration. Oncotarget. 2016;7(2):1584–1597. doi:10.18632/oncotarget.6444
15. Krupp M, Weinmann A, Galle PR, Teufel A. Actin binding LIM protein 3 (abLIM3). Int J Mol Med. 2006;17(1):129–133.
16. Soon PS, Gill AJ, Benn DE, et al. Microarray gene expression and immunohistochemistry analyses of adrenocortical tumors identify IGF2 and Ki-67 as useful in differentiating carcinomas from adenomas. Endocr Relat Cancer. 2009;16(2):573–583. doi:10.1677/ERC-08-0237
17. Li YJ, Yang Z, Wang YY, Wang Y. Long noncoding RNA ZNF667-AS1 reduces tumor invasion and metastasis in cervical cancer by counteracting microRNA-93-3p-dependent PEG3 downregulation. Mol Oncol. 2019;13(11):2375–2392. doi:10.1002/1878-0261.12565
18. Meng W, Cui W, Zhao L, Chi W, Cao H, Wang B. Aberrant methylation and downregulation of ZNF667-AS1 and ZNF667 promote the malignant progression of laryngeal squamous cell carcinoma. J Biomed Sci. 2019;26(1):13. doi:10.1186/s12929-019-0506-0
19. Zheng ZQ, Li ZX, Zhou GQ, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79(18):4612–4626. doi:10.1158/0008-5472.CAN-19-0799
20. Herold C, Hooli BV, Mullin K, et al. Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer’s disease with OSBPL6, PTPRG, and PDCL3. Mol Psychiatry. 2016;21(11):1608–1612. doi:10.1038/mp.2015.218
21. Chen J, Jiang C, Fu L, et al. CHL1 suppresses tumor growth and metastasis in nasopharyngeal carcinoma by repressing PI3K/AKT signaling pathway via interaction with Integrin beta1 and Merlin. Int J Biol Sci. 2019;15(9):1802–1815. doi:10.7150/ijbs.34785
22. Xue K, Cao J, Wang Y, et al. Identification of potential therapeutic gene markers in nasopharyngeal carcinoma based on bioinformatics analysis. Clin Transl Sci. 2019.
23. Zhang W, Wang L, Zheng F, et al. Long noncoding RNA expression signatures of metastatic nasopharyngeal carcinoma and their prognostic value. Biomed Res Int. 2015;2015:618924. doi:10.1155/2015/618924
24. Jin JJ, Liu YH, Si JM, Ni R, Wang J. Overexpression of miR-1290 contributes to cell proliferation and invasion of non small cell lung cancer by targeting interferon regulatory factor 2. Int J Biochem Cell Biol. 2018;95:113–120. doi:10.1016/j.biocel.2017.12.017
25. Zhao LP, Li RH, Han DM, et al. Independent prognostic factor of low-expressed LncRNA ZNF667-AS1 for cervical cancer and inhibitory function on the proliferation of cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21(23):5353–5360. doi:10.26355/eurrev_201712_13920
26. Li JW, Kuang Y, Chen L, Wang JF. LncRNA ZNF667-AS1 inhibits inflammatory response and promotes recovery of spinal cord injury via suppressing JAK-STAT pathway. Eur Rev Med Pharmacol Sci. 2018;22(22):7614–7620. doi:10.26355/eurrev_201811_16375
27. Lian Y, Xiong F, Yang L, et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37(1):253. doi:10.1186/s13046-018-0918-9
28. Gong Z, Yang Q, Zeng Z, et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37(3):3683–3695. doi:10.1007/s13277-015-4156-x
29. Sun H, Wang L, Zhao Q, Dai J. Diagnostic and prognostic value of serum miRNA-1290 in human esophageal squamous cell carcinoma. Cancer Biomark. 2019;25(4):381–387. doi:10.3233/CBM-190007
30. Mao Y, Liu J, Zhang D, Li B. MiR-1290 promotes cancer progression by targeting nuclear factor I/X(NFIX) in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother. 2015;76:82–93. doi:10.1016/j.biopha.2015.10.005
31. Xie R, Wu SN, Gao CC, et al. Prognostic value of combined and individual expression of microRNA-1290 and its target gene nuclear factor I/X in human esophageal squamous cell carcinoma. Cancer Biomark. 2017;20(3):325–331. doi:10.3233/CBM-170029
32. Janiszewska J, Szaumkessel M, Kostrzewska-Poczekaj M, et al. Global miRNA expression profiling identifies miR-1290 as novel potential oncomiR in laryngeal carcinoma. PLoS One. 2015;10(12):e0144924. doi:10.1371/journal.pone.0144924
33. Jin SH, Kim H, Gu DR, et al. Actin-binding LIM protein 1 regulates receptor activator of NF-kappaB ligand-mediated osteoclast differentiation and motility. BMB Rep. 2018;51(7):356–361. doi:10.5483/BMBRep.2018.51.7.106
34. Kwong LN, Chromosome CL. 10, frequently lost in human melanoma, encodes multiple tumor-suppressive functions. Cancer Res. 2014;74(6):1814–1821. doi:10.1158/0008-5472.CAN-13-1446
35. Narahara H, Sakai E, Yamaguchi Y, et al. Actin binding LIM 1 (abLIM1) negatively controls osteoclastogenesis by regulating cell migration and fusion. J Cell Physiol. 2018;234(1):486–499. doi:10.1002/jcp.26605
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.