Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA TTN-AS1 Regulates miR-524-5p and RRM2 to Promote Breast Cancer Progression
Authors Feng H, Wang Q, Xiao W, Zhang B, Jin Y, Lu H
Received 23 December 2019
Accepted for publication 7 April 2020
Published 27 May 2020 Volume 2020:13 Pages 4799—4811
DOI https://doi.org/10.2147/OTT.S243482
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Hui Feng, Qi Wang, Wenjing Xiao, Biyuan Zhang, Yonglong Jin, Haijun Lu
Department of Radiotherapy, Affiliated Hospital of Qingdao University, Qingdao 266000, Shandong, People’s Republic of China
Correspondence: Haijun Lu
Department of Radiotherapy, Affiliated Hospital of Qingdao University, Jiangsu Road No. 16, Shinan District, Qingdao 266000, Shandong, People’s Republic of China
Tel +8618661809125
Email [email protected]
Background: Recent studies suggest many long non-coding RNAs (lncRNAs) are crucial oncogenes or tumor suppressors. This study intended to investigate the biological function and mechanism of lncRNA TTN antisense RNA 1 (TTN-AS1) in the progression of breast cancer (BC).
Materials and Methods: BC tissue samples were collected. The expression of TTN-AS1 in BC tissues and adjacent tissues was detected by qRT-PCR, and the relationship between pathological indicators and TTN-AS1 expression was analyzed by chi-square test. BC cell lines T47D and BT549 were utilized as cell models. CCK-8 assay and BrdU assay were used to detect the effect of TTN-AS1 on BC cell proliferation. Transwell assay was used to detect the effects of TTN-AS1 on cell migration and invasion. In addition, dual-luciferase reporter gene assay was used to confirm the targeting relationship between miR-524-5p and TTN-AS1. Western blot was used to detect the function of TTN-AS1 on regulating ribonucleotide reductase subunit 2 (RRM2) and survivin. Additionally, subcutaneous xenotransplanted tumor model and tail vein injection model were constructed in vivo.
Results: The expression of TTN-AS1 in BC tissues was significantly higher than that in normal tissues, and its high expression was correlated with adverse pathological indicators. Overexpression of TTN-AS1 significantly promoted the proliferation, migration and invasion of BC cells. TTN-AS1 knockdown suppressed the malignant phenotypes of BC cells. TTN-AS1 overexpression significantly impeded the expression of miR-524-5p, but increased the expression of RRM2.
Conclusion: TTN-AS1 exerts oncogenic function in BC by repressing miR-524-5p and increasing the expression of RRM2.
Keywords: breast cancer, lncRNA, TTN-AS1, miR-524-5p, RRM2
Introduction
Breast cancer (BC) is one of the most common cancers, and one of the leading causes of cancer-related deaths in women.1,2 The early diagnosis rate of BC cases is low, and most of the patients are already in advanced stages when diagnosed.3 At present, surgery is still the main method to treat BC. Meanwhile, with the application of chemotherapy, radiotherapy and endocrine therapy, the survival rate of BC patients has been significantly improved. However, there are still a considerable number of patients suffering from recurrence or drug resistance.4,5 Therefore, it is of great significance to investigate the molecular mechanism of BC progression to look for novel therapy targets.
Long non-coding RNA (LncRNA) is a class of non-coding RNAs longer than 200 nt. They do not encode proteins, but they regulate gene expression at different levels such as epigenetic regulation, transcriptional regulation and post-transcriptional regulation.6,7 Recent studies have indicated that lncRNAs exert a crucial regulatory role in a variety of malignancies.8,9 TTN-AS1 is a lncRNA which is involved in the progression of cancers such as colorectal cancer, prostate cancer and osteosarcoma.10–12 Instead, whether TTN-AS1 participates in BC progression awaits further investigations.
MiRNA is a class of short non-coding RNAs with a length of about 20 nucleotides and highly conserved sequence. It regulates gene expression by binding with the 3ʹ-untranslated region (3ʹUTR) of mRNA.13 Recent studies have proved that miRNAs participate in regulating cancer cell proliferation, apoptosis, migration and invasion.14,15 miR-524-5p has been widely studied in cancer biology. It is down-regulated in gastric cancer tissues, and inhibits gastric cancer cell proliferation and promotes apoptosis via modulating CASP3;16 it also inhibits the progression of papillary thyroid carcinoma cells via targeting FOXE1 and ITGA3.17 However, the expression pattern and function of miR-524-5p in BC are unclear.
Ribonucleotide reductase regulatory subunit M2 (RRM2) is an important component of ribonucleotide reductase (RNR), which is the crucial enzyme in the de novo synthesis of deoxyribonucleotide triphosphates (dNTPs).18 RRM2 exerts an essential role in regulating RNR enzyme activity, DNA synthesis and cell proliferation, and is considered as a potential anti-cancer therapeutic target.18 Accumulating researches prove that RRM2 is highly expressed in prostate cancer, cervical cancer, and BC, and modulates biological behaviors like cancer cell proliferation, differentiation, metastasis, and drug resistance.19–21 However, further investigation on RRM2 is required to fully reveal the mechanism of its dysregulation in cancer.
The purpose of this study was to investigate the expression, clinical significance and mechanism of TTN-AS1 in BC. The present study confirmed that TTN-AS1 promoted BC progression by targeting miR-524-5p to up-regulate RRM2. This work revealed a new molecular mechanism in the progression of BC, and provided a new theoretical basis for the treatments of BC.
Materials and Methods
Tissue Samples
This study was approved by the Ethics Review Board of Affiliated Hospital of Qingdao University. Fifty-six cases of BC specimens and the corresponding adjacent normal tissues were randomly collected. These tissues were surgically resected in Affiliated Hospital of Qingdao University from September 2016 to May 2018. The patients enrolled signed the informed consent. All patients did not receive radiotherapy or chemotherapy before surgery. The tissue samples were immediately stored in liquid nitrogen at −196°C after removal for subsequent experiments.
Cell Culture
The cells used in this study were purchased from China Center for Type Culture Collection (Wuhan, China). Human normal mammary epithelial cell line MCF-10A cells were cultured in DMEM (Thermo Fisher Scientific, Shanghai, China), and BC cell lines (T47D, BT549, SKBR3, and BT474) were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Shanghai, China). 10% fetal bovine serum (FBS; Thermo Fisher Scientific, MA, USA), 100 U/mL penicillin (Hyclone, Logan, UT, USA), and 0.1 mg/mL streptomycin (Hyclone, Logan, UT, USA) were added into the medium, and the cells were cultured at 37°C in an incubator in a saturated humidity atmosphere with 5% CO2. When the cell confluence reaches 70% to 90%, 0.25% trypsin (Hyclone, Logan, UT, USA) was used for passage.
Cell Transfection
BC cells in logarithmic growth phase were trypsinized, harvested and resuspended, and the cell concentration was adjusted to 1 × 105 cells/mL. The cells were inoculated into a 6-well plate, and 2 mL of cell suspension was loaded into each well, and the cells were cultured for 24 h. pcDNA-TTN-AS1, TTN-AS1 shRNA, miR-524-5p mimics and inhibitors were transfected into BC cells in compliance with the instructions of Lipofectamine 2000 transfection reagent (Invitrogen, Shanghai, China); 48 h after transfection, the transfection efficiency was detected by quantitative real-time polymerase chain reaction (qRT-PCR).
qRT-PCR
The total RNA of BC tissues or cells were extracted in accordance with the instructions of TRIzol reagent (Thermo Fisher Scientific, Shanghai, China). NanoDrop Microvolume Spectrophotometers and Fluorometer (Thermo Fisher Scientific, Shanghai, China) was used to detect the concentration and purity of RNA and then cDNA was synthesized by reverse transcription with Reverse Transcription Kit (Takara, Dalian, China). After that, PCR reaction system with a final volume of 20 μL was established: 2 μL of cDNA, 10 μL of SYBR Premix Ex Taq™ Kit (Takara, Dalian, China), 1 μL of upstream and downstream primers (10 μmol/L), and 7 μL ddH2O. The parameters of PCR thermal cycle are: 95°C for 5 min, then 3 steps were followed by: denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 60 s, 45 cycles. The primer sequences were as follows: TTN-AS1: F: 5ʹ-CGGGAACAAGCCCTGTG-3ʹ, R: 5ʹ-CCGGCCCAAAGATGATG-3ʹ; GAPDH: F: 5ʹ-TGCACCACCACCTGCTTAGC-3ʹ, R: 5ʹ-GGCATGGACTGTGGTCATGAG-3ʹ; miR-524-5p: F: 5ʹ-CTACAAAGGGAAGCACTTTTCTCAA-3ʹ, R: Uni-miR qPCR primer; U6: F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, R: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. The relative expression of the genes was calculated using 2− ΔΔ Ct method.
Cell Proliferation Assay
BC cells in the logarithmic growth phase were trypsinized, harvested and resuspended and then the cell suspension was adjusted to a concentration of 1 × 104/mL. 100 μL of cell suspension was dripped into each well of a 96-well plate. After the cells were cultured for 24 h, 48 h, 72 h and 96 h after inoculation, 10 μL of CCK-8 reagent was loaded (Beyotime, Beijing, China) to each well. After CCK-8 reagent was added, the culture was continued for 1 h, and then the optical density (OD) values at 450 nm of the cells were measured with a microplate reader.
BrdU Staining
BC cells were inoculated onto coverslips placed in 24-well plate. 200 μL BrdU staining reagent (Beyotime, Beijing, China) was dripped into each well, and cells were incubated in the incubator for 2 h and then rinsed with PBS. After that, the cells were fixed with paraformaldehyde and incubated for 10 min at room temperature. Then, 200 μL glycine at a concentration of 2 mg/mL was added, and the cells were incubated for 5 min, and washed with PBS on the shaking table for 5 min. After that, 100 μL PBS containing 0.5% TritonX-100 was dripped into each well, and cells were placed on the shaking table and washed twice with PBS for 5 min each time. Then, the cells were stained with fluorescent dye Apollo (Beyotime, Beijing, China) at room temperature for 30 min, and then incubated with 1 × Hoechst 33342 DNA-staining solution (Beyotime, Beijing, China) in dark at room temperature for 20 min. Then, the cells were rinsed with PBS, observed, photographed and counted under a fluorescence microscope.
Transwell Experiment
In migration assay, 1 × 105 cells resuspended in 200 μL serum-free medium were inoculated in the Transwell Chambers (8 μm pore diameter, BD Biosciences, CA, USA) placed in a 24-well plate, and 700 μL medium containing 20% FBS was dripped into the wells of 24-well plate. Then, the 24-well plate was placed in an incubator for 36 h. Next, the medium was discarded, and the Transwell chambers were removed, and cells that did not pass through the membrane were gently wiped off with a wet cotton swab. The migrated cells were fixed with methanol for 30 s, stained with crystal violet for 30 s, then washed with PBS. After that, the cells were observed under a microscope and the cells in six visual fields were randomly selected and counted. The invasion experiment used Transwell chamber which was covered with Matrigel (Millipore, Billerica, MA, USA), and the other steps were the same as the cell migration experiment.
Dual-Luciferase Reporter Gene Assay
The binding sequences between TTN-AS1 or RRM2 and miR-524-5p were amplified by PCR, respectively. The amplified product was inserted into the pmirGLO dual-luciferase target expression vector to construct TTN-AS1 and RRM2 wild-type (WT) plasmids; TTN-AS1 and RRM2 mutant (Mut) plasmids were constructed by site-directed mutagenesis (Thermo Fisher Scientific, Shanghai, China). The recombinant plasmids were co-transfected into HEK293T cells with miR-524-5p or miR-NC, respectively; 48 h after transfection, cells were harvested. Then, the luciferase activity was measured using the Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA).
Western Blot Assay
Cells were rinsed with pre-cooled PBS 2 to 3 times, and then RIPA lysate (containing 1% PMSF) was added to extract the total protein. The protein concentration was determined by BCA assay. After denaturation, equal amount of protein samples were separated by 10% SDS-PAGE. Then, the protein was transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA) and blocked with 5% skim milk for 1 h. After incubated with primary antibodies anti-RRM2 (ab57653, Abcam, 1:1000), anti-Survivin antibody (ab76424, Abcam, 1:1000) and anti-GAPDH (ab8245, Abcam, 1:1000) at 4°C overnight, the membrane was reacted with the secondary antibody (1:5000, Biogot Technology, Nanjing, China). After that, signals were visualized using an enhanced chemiluminescence detection kit (Beyotime, Beijing, China).
In vivo Tumorigenesis Experiment
All animal experiments were backed by the Affiliated Hospital of Qingdao University Animal Care and Use Committee. The protocols of the animal experiments in this study followed the United Kingdom Coordinating Committee on Cancer Research (UKCCCR) guidelines for the welfare of animals in experimental neoplasia. Female athymic BALB/c nude mice (4 weeks old) were purchased from Wuxi Biomedical Technology Co., Ltd. (Wuxi, China). BT549 cells were divided into control (sh-NC) group and TTN-AS1 knockdown (sh-TTN-AS1) group. 1 × 107 cells in each group were subcutaneously injected into the back of each mouse (left: sh-TTN-AS1; right: sh-NC), and the tumor volume was measured every 7 days. Mice were sacrificed on day 35 after injection, and xenograft tumors were excised and weighed. In the lung metastasis study, there were 5 mice in each group. 1×107 cells of each group were injected into the caudal vein of the mice. Three weeks later, the mice were sacrificed and the lungs were removed and fixed in formaldehyde, and then embedded in paraffin. At last, pathological examination was used to quantify lung metastasis.
Statistical Analysis
Graphpad Prism 7.0 software (Graphpad, Inc., La Jolla, CA, USA) was adopted for statistical processing. All data were expressed as mean ± SD. χ2 test was used to evaluate the relationship between TTN-AS1 expression and the clinicopathological characteristics of tumors, and correlation between two genes was performed using Pearson’s correlation analysis. Differences of P < 0.05 were considered statistically significant.
Results
TTN-AS1 Was Up-Regulated in BC Samples, Which Was Related to the Pathological Parameters of the Patients
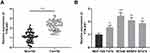
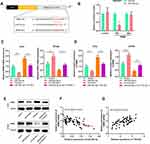
Firstly, we detected the expression of TTN-AS1 in 56 BC samples and adjacent tissue samples. Compared with adjacent normal tissues, TTN-AS1 was expressed at a higher level in BC tissues (Figure 1A). Moreover, compared with normal breast cell line MCF-10A, TTN-AS1 expression was higher in BC cell lines (Figure 1B). Next, the aforementioned 56 BC samples were used to analyze the correlation between TTN-AS1 expression and tumor pathological parameters in patients with BC (Table 1). Chi-square test showed that high expression of TTN-AS1 in tumor tissues was closely related to larger tumor size (P = 0.0130), local lymph node invasion (P = 0.0042) and higher TNM stage (P = 0.0010) in BC patients, suggesting that TTN-AS1 could probably promote the occurrence and metastasis of BC.
 |
Table 1 Correlation Between Clinicopathological Indicators and TTN-AS1 Expression in 56 BC Patients |
TTN-AS1 Could Promote the Proliferation, Migration and Invasion of BC Cells
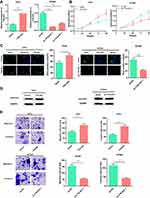
Next, the function of TTN-AS1 in BC cells was explored. Based on expression of TTN-AS1 in the four BC cells, we selected T47D and BT549 cell lines to successfully construct a TTN-AS1 overexpression model and a TTN-AS1 knockdown model, respectively (Figure 2A). On this basis, CCK-8 assay was used to detect the proliferation ability of BC cells. The results suggested that compared with the control group in BT549 cells, the proliferation ability of TTN-AS1 knockdown group was significantly inhibited; in contrast, TTN-AS1 over-expression promoted the proliferation of T47D cells (Figure 2B). Besides, the proliferation of BC cells was further detected using BrdU assay. The results manifested that the number of BrdU-positive cells in the TTN-AS1 knockdown group was significantly reduced in BT549 cells, while over-expression of TTN-AS1 increased the number of BrdU-positive cells in T47D cells (Figure 2C). Next, Western blot was used to detect the expression of apoptosis-inhibiting protein Survivin. As shown, over-expression of TTN-AS1 promoted Survivin expression, while knockdown of TTN-AS1 reduced Survivin expression (Figure 2D). Additionally, the effect of TTN-AS1 on cell migration and invasion was evaluated through Transwell assay. The results demonstrated that compared with the control groups, the number of migration and invasion of BT549 cells with TTN-AS1 knockdown was decreased significantly; TTN-AS1 overexpression significantly facilitated the migration and invasion of T47D cells (Figure 2E). Collectively, these results indicated that TTN-AS1 could promote the malignant phenotypes of BC cells.
TTN-AS1 Was a Molecular Sponge for miR-524-5p
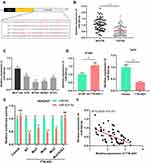
To clarify the downstream mechanism of TTN-AS1 regulating BC progression, we performed a bioinformatics analysis through StarBase database (starbase.sysu.edu.cn). It was implied that TTN-AS1 contained 3 binding sites for miR-524 −5p (Figure 3A). Compared with normal tissues and cells, miR-524-5p was down-regulated in cancer tissues and cells (Figure 3B and C). To further verify whether miR-524-5p was indeed a target of TTN-AS1, after TTN-AS1 was overexpressed or knocked down in BC cells, the expression of miR-524-5p was detected by qRT-PCR. It was observed that miR-524-5p expression was reduced in TTN-AS1-overexpressing cells, while increased in TTN-AS1 knockdown cells (Figure 3D). Additionally, dual-luciferase reporter assay indicated that miR-524-5p reduced the luciferase activity of wild-type TTN-AS1 reporters, but had no significant effect on the reporter when all of the 3 predicted binding sites were mutated (Figure 3E). Additionally, the correlation between the expressions of TTN-AS1 and miR-524-5p in BC tissues was analyzed. The results manifested that the expression of TTN-AS1 in BC samples was negatively correlated with the expression of miR-524-5p (Figure 3F). These results indicated that miR-524-5p could directly bind to TTN-AS1 at the miRNA recognition site, and was negatively regulated by TTN-AS1.
miR-524-5p Reversed the Cancer-Promoting Effect Caused by Over-Expression of TTN-AS1
To verify the function of TTN-AS1/miR-524-5p axis in BC progression, we transfected miR-524-5p inhibitors and miR-524-5p mimics into BT549 cells with over-expressed TTN-AS1 and T47D cells with TTN-AS1 knockdown, respectively (Figure 4A). We found that miR-524-5p over-expression attenuated the effects of TTN-AS1 overexpression on BC cell proliferation, migration, and invasion; consistently, miR-524-5p inhibition reversed the effects induced by TTN-AS1 knockdown (Figure 4B–D). These findings suggested that TTN-AS1 promoted BC cell proliferation, migration, and invasion through repressing miR-524-5p.
TTN-AS1/miR-524-5p Axis Regulated RRM2 Expression
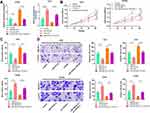
After confirming that TTN-AS1 can regulate miR-524-5p expression, we tried to explore the downstream targets of miR-524-5p. Through TargetScan database (www.targetscan.org/), it was found that the 3ʹUTR of RRM2 contained a potential target site for miR-524-5p (Figure 5A). Next, dual-luciferase reporter gene assay confirmed that miR-524-5p could bind with the wile type 3ʹUTR of RRM2, but could not bind with the mutant 3ʹUTR of RRM2 (Figure 5B). Subsequently, it was investigated whether TTN-AS1 could regulate RRM2 expression, and it was found that TTN-AS1 knockdown could considerably reduce the mRNA and protein expression levels of RRM2, and TTN-AS1 overexpression functioned oppositely; in addition, transfection of miR-524-5p mimics reduced RRM2 expression and attenuated the effect of TTN-AS1-induced up-regulation of RRM2; inhibition of miR-524-5p increased the RRM2 expression and reversed the effect of TTN-AS1 knockdown on RRM2 expression (Figure 5C–E). Importantly it was demonstrated that the expression of miR-524-5p was negatively correlated with the expression of RRM2 in BC samples, while the expression of TTN-AS1 was positively correlated with the expression of RRM2 in BC samples (Figure 5F and G). These results indicated that TTN-AS1 could regulate RRM2 expression in BC cells by modulating miR-524-5p.
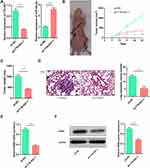
TTN-AS1 Regulated the Growth and Metastasis of BC Cells in vivo
To investigate whether TTN-AS1 could affect tumor growth and metastasis in vivo, lentiviral vector was adopted to construct stable BT549 cell line with TTN-AS1 knocked down (Figure 6A). Based on this, a subcutaneous tumorigenesis model was established. It was demonstrated that the size of the tumor formed by BT549/sh-TTN-AS1 cells was significantly smaller than that formed by BT549/sh-NC cells (Figure 6B). Consistently, it was found that the weight of tumor formed by BT549/sh-TTN-AS1 cells was significantly less than that of BT549/sh-NC cells (Figure 6C). Additionally, BT549/sh-TTN-AS1 and BT549/sh-NC cells were transplanted into the caudal vein of nude mice to further determine the effect of TTN-AS1 knockdown on tumor metastasis in vivo. As shown, compared with BT549/sh-NC group, the number of lung metastasis in mice injected with BT549/sh-TTN-AS1 cells was significantly reduced (Figure 6D). qRT-PCR and Western blot were adopted to detect the expression of RRM2 in the above tumors and it was found that the RRM2 in the sh-TTN-AS1 group was reduced than that in the sh-NC group (Figure 6E and F). These results indicated that knocking down TTN-AS1 could inhibit the growth and metastasis of BC cells in vivo via regulating miR-524-5p and RRM2.
Discussion
LncRNA was initially considered to be the “noise” of genome transcription without biological function. However, in recent years, researches have found that lncRNA can modulate the expression of coding genes at different levels, thereby participating in a variety of important biological processes, like species evolution, embryonic development, maintenance of stem cells, cell proliferation, differentiation, tumorigenesis, cancer progression and so on.22,23 Multiple studies have demonstrated that lncRNA is abnormally expressed, and plays an important role in the occurrence, development and metastasis of BC.24,25 For example, lncRNA LUCAT1 is highly expressed in BC, and over-expression of LUCAT1 promotes the proliferation, migration and invasion of BC cells.24 LncRNA SNHG3 is up-regulated in BC and promotes the proliferation and invasion of BC cells via regulating the miR-384/HDGF axis.25 Previous studies have shown that TTN-AS1 exerts an essential role in a variety of tumors. For example, TTN-AS1 regulates CDK5 through adsorbing miR-142-5p, and promotes the migration, invasion, and epithelial-mesenchymal transition of lung adenocarcinoma cells.26 In cervical cancer, TTN-AS1 promotes cancer progression by regulating miR-573/E2F3 axis.27 In the current study, it was demonstrated for the first time that TTN-AS1 was up-regulated in BC tissues and cells, and its high expression was significantly correlated with poor clinicopathological indicators. It was also demonstrated that TTN-AS1 over-expression promoted cell proliferation, migration and invasion, and knocking down TTN-AS1 had opposite effects. Our in vitro and in vivo data confirmed that TTN-AS1 played an oncogenic role in BC progression.
As an endogenous non-coding RNA, miRNA is one of the crucial factors that is at play in the tumorigenesis and progression of BC. It participated in many biological processes including cell proliferation, differentiation, apoptosis, migration and invasion. For example, miR-425-5p is associated with poor prognosis in BC patients and promotes tumor cell progression via targeting PTEN;28 miR-216b-5p inhibits human BC cell proliferation by down-regulating HDAC8 expression;29 miR-21 promotes the proliferation and metastasis of BC cells via targeting LZTFL1.30 miR-524-5p is involved in the progression of gastric cancer, papillary thyroid carcinoma, melanoma, glioma and pituitary adenoma.16,17,31–34 In this work, it was observed that the expression of miR-524-5p was significantly down-regulated in BC tissues, suggesting that it functioned as a tumor suppressor. After TTN-AS1 was overexpressed or knocked down, qRT-PCR was used to detect miR-524-5p expression, and the results implied that TTN-AS1 negatively regulated miR-524-5p expression in BC cells. Consistently, in BC tissues, TTN-AS1 expression was negatively correlated with miR-524-5p expression. Additionally, it was confirmed that there was a targeted binding relationship between miR-524-5p and TTN-AS1 by dual-luciferase reporter assay. These results in the present work indicated that TTN-AS1 functioned as an upstream molecule of miR-524-5p in BC, and contributed to its dysregulation.
RNR features prominently in DNA synthesis, DNA damage repair and cell proliferation. RNR is composed of three subunits, namely the large subunit RRM1, the small subunit RRM2, and the small subunit p53R2. Abnormal expression of RRM2 is found in human cancers such as gastric cancer, adrenocortical cancer, pancreatic cancer and bladder cancer. Inhibition of RRM2 can reduce the activity of RR enzymes, restrain the growth of cancer cells, facilitate cancer cell apoptosis, impede tumor metastasis and reverse drug resistance.35–38 In this study, RRM2 was identified as a target gene of miR-524-5p, and it was demonstrated that miR-524-5p could negatively regulate RRM2 expression. Additionally, it was found that TTN-AS1 and RRM2 expressions were positively correlated in BC tissues. And in vitro experiments also found that overexpression of TTN-AS1 can promote RRM2 expression. Next, through rescue experiments, it was proved that miR-524-5p could partially reverse the promotion effect of TTN-AS1 on RRM2 expression, thereby affecting cell proliferation and metastasis. Based on the above data, it was concluded that TTN-AS1 could promote the expression of RRM2 through targetedly regulating miR-524-5p, promoting the occurrence and progression of BC cells.
In summary, our study for the first time confirms that TTN-AS1 is highly expressed in BC, and its high expression is significantly related to the poor prognosis of patients. In addition, our research also demonstrates that TTN-AS1 can up-regulate RRM2 expression through modulating miR-524-5p, thereby promoting BC cell proliferation and metastasis. Our work depicts a new molecular mechanism in the progression of BC, namely TTN-AS1/miR-524-5p/RRM2 axis, and provides potential novel biomarkers and therapy targets for BC.
Data Sharing Statement
All data included in this study are available upon request by contact with the corresponding author.
Ethics and Consent Statement
Our study was approved by the Ethics Review Board of Affiliated Hospital of Qingdao University.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
1. Coleman C. Early detection and screening for breast cancer. Semin Oncol Nurs. 2017;33(2):141–155. doi:10.1016/j.soncn.2017.02.009
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
3. Weigelt B, Peterse JL, van ‘T Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi:10.1038/nrc1670
4. Kutanzi KR, Yurchenko OV, Beland FA, Checkhun VF, Pogribny IP. MicroRNA-mediated drug resistance in breast cancer. Clin Epigenetics. 2011;2(2):171–185. doi:10.1007/s13148-011-0040-8
5. Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–234. doi:10.1038/nrd1984
6. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606
7. Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi:10.1073/pnas.0904715106
8. Sun M
9. Joung J, Engreitz JM, Konermann S, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548(7667):343–346. doi:10.1038/nature23451
10. Wang Y, Jiang F, Xiong Y, Cheng X, Qiu Z, Song R. LncRNA TTN-AS1 sponges miR-376a-3p to promote colorectal cancer progression via upregulating KLF15. Life Sci. 2019;11:116936.
11. Luo JF, Xu J, Zheng JZ. Long non-coding RNA TTN-AS1 promotes cell proliferation and inhibits cell apoptosis in prostatic cancer by sponging miR-193a-5p. Eur Rev Med Pharmacol Sci. 2019;23(18):7816–7825. doi:10.26355/eurrev_201909_18991
12. Fu D, Lu C, Qu X, et al. LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY). 2019;11(19):8374–8385. doi:10.18632/aging.102325
13. Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi:10.1016/j.cell.2009.01.046
14. Ma Y, Ma M, Ma L, Zhang F, Liu Y, Ma X. Downregulation of miR-552 in hepatocellular carcinoma inhibits cell migration and invasion, and promotes cell apoptosis via RUNX3. Exp Ther Med. 2019;18(5):3829–3836. doi:10.3892/etm.2019.8061
15. Mo JS, Park WC, Choi SC, Yun KJ, Chae SC. MicroRNA 452 regulates cell proliferation, cell migration, and angiogenesis in colorectal cancer by suppressing VEGFA expression. Cancers (Basel). 2019;11(10):E1613. doi:10.3390/cancers11101613
16. Zhu C-Y, Meng F-Q, Liu J. MicroRNA-524-5p suppresses cell proliferation and promotes cell apoptosis in gastric cancer by regulating CASP3. Eur Rev Med Pharmacol Sci. 2019;23(18):7968–7977. doi:10.26355/eurrev_201909_19013
17. Liu H, Chen X, Lin T, Chen X, Yan J, Jiang S. MicroRNA-524-5p suppresses the progression of papillary thyroid carcinoma cells via targeting on FOXE1 and ITGA3 in cell autophagy and cycling pathways. J Cell Physiol. 2019;234(10):18382–18391. doi:10.1002/jcp.28472
18. Le Calvé B, Rynkowski M, Le Mercier M, et al. Long-term in vitro treatment of human glioblastoma cells with temozolomide increases resistance in vivo through up-regulation of GLUT transporter and aldo-keto reductase enzyme AKR1C expression. Neoplasia. 2010;12(9):727–739. doi:10.1593/neo.10526
19. Gan BL, Zhang LJ, Gao L, et al. Downregulation of miR-224-5p in prostate cancer and its relevant molecular mechanism via TCGA, GEO database and in silico analyses. Oncol Rep. 2018;40(6):3171–3188.
20. Wang N, Zhan T, Ke T, et al. Increased expression of RRM2 by human papillomavirus E7 oncoprotein promotes angiogenesis in cervical cancer. Br J Cancer. 2014;110(4):1034–1044. doi:10.1038/bjc.2013.817
21. Liang WH, Li N, Yuan ZQ, Qian XL, Wang ZH. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol Carcinog. 2019;58(4):461–473. doi:10.1002/mc.22941
22. Shih JW, Chiang WF, Wu ATH, et al. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1α co-activator driving oral cancer progression. Nat Commun. 2017;8:15874. doi:10.1038/ncomms15874
23. Liu T, Zhang X, Yang YM, Du LT, Wang CX. Increased expression of the long noncoding RNA CRNDE-h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther. 2016;9:1437–1448. doi:10.2147/OTT.S98268
24. Li YL, Wang XM, Qiao GD, et al. Up-regulated lnc-lung cancer associated transcript 1 enhances cell migration and invasion in breast cancer progression. Biochem Biophys Res Commun. 2019;
25. Ma Q, Qi X, Lin X, Li L, Chen L, Hu W. LncRNA SNHG3 promotes cell proliferation and invasion through the miR-384/hepatoma-derived growth factor axis in breast cancer. Hum Cell. 2020;33(1):232–242.
26. Jia Y, Duan Y, Liu T, et al. LncRNA TTN-AS1 promotes migration, invasion, and epithelial mesenchymal transition of lung adenocarcinoma via sponging miR-142-5p to regulate CDK5. Cell Death Dis. 2019;10(8):573. doi:10.1038/s41419-019-1811-y
27. Chen P, Wang R, Yue Q, Hao M. Long non-coding RNA TTN-AS1 promotes cell growth and metastasis in cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun. 2018;503(4):2956–2962. doi:10.1016/j.bbrc.2018.08.077
28. Xiao S, Zhu H, Luo J, Wu Z, Xie M. miR‑425‑5p is associated with poor prognosis in patients with breast cancer and promotes cancer cell progression by targeting PTEN. Oncol Rep. 2019. doi:10.3892/or.2019.7371
29. Menbari MN, Rahimi K, Ahmadi A, et al. MiR-216b-5p inhibits cell proliferation in human breast cancer by down-regulating HDAC8 expression. Life Sci. 2019;237:116945. doi:10.1016/j.lfs.2019.116945
30. Wang H, Tan Z, Hu H, et al. microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer. 2019;19(1):738. doi:10.1186/s12885-019-5951-3
31. Wang J, Xue X, Hong H, et al. Upregulation of microRNA-524-5p enhances the cisplatin sensitivity of gastric cancer cells by modulating proliferation and metastasis via targeting SOX9. Oncotarget. 2017;8(1):574–582. doi:10.18632/oncotarget.13479
32. Liu SM, Lu J, Lee HC, Chung FH, Ma N. miR-524-5p suppresses the growth of oncogenic BRAF melanoma by targeting BRAF and ERK2. Oncotarget. 2014;5(19):9444–9459. doi:10.18632/oncotarget.2452
33. Chen L, Zhang W, Yan W, et al. The putative tumor suppressor miR-524-5p directly targets Jagged-1 and Hes-1 in glioma. Carcinogenesis. 2012;33(11):2276–2282. doi:10.1093/carcin/bgs261
34. Zhen W, Qiu D, Zhiyong C, et al. MicroRNA-524-5p functions as a tumor suppressor in a human pituitary tumor-derived cell line. Horm Metab Res. 2017;49(7):550–557. doi:10.1055/s-0043-106437
35. Zhong Z, Cao Y, Yang S, Zhang S. Overexpression of RRM2 in gastric cancer cell promotes their invasiveness via AKT/NF-κB signaling pathway. Pharmazie. 2016;71(5):280–284.
36. Grolmusz VK, Karászi K, Micsik T, et al. Cell cycle dependent RRM2 may serve as proliferation marker and pharmaceutical target in adrenocortical cancer. Am J Cancer Res. 2016;6(9):2041–2053.
37. Xia G, Wang H, Song Z, Meng Q, Huang X, Huang X. Gambogic acid sensitizes gemcitabine efficacy in pancreatic cancer by reducing the expression of ribonucleotide reductase subunit-M2 (RRM2). J Exp Clin Cancer Res. 2017;36(1):107. doi:10.1186/s13046-017-0579-0
38. Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology. 2010;57(6):885–892. doi:10.1111/j.1365-2559.2010.03725.x
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.