Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA TTN-AS1 Promotes Progression of Non-Small Cell Lung Cancer via Regulating miR-491-5p/ZNF503 Axis
Received 16 November 2019
Accepted for publication 19 March 2020
Published 1 July 2020 Volume 2020:13 Pages 6361—6371
DOI https://doi.org/10.2147/OTT.S238890
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Guanbin Qi, Lei Li
Department of Respiratory and Critical Care Medicine, Huaihe Hospital, Henan University, Kaifeng, Henan 475000, People’s Republic of China
Correspondence: Lei Li
Department of Respiratory and Critical Care Medicine, Huaihe Hospital, Henan University, No. 115, Ximen Street, Kaifeng City, Henan Province 475000, People’s Republic of China
Tel +86-371-23906701
Email [email protected]
Background: Non-small cell lung cancer (NSCLC) is the most common type of lung cancer with high mortality worldwide. Long non-coding RNA (lncRNA) TTN antisense RNA1 (TTN-AS1) has been demonstrated to play a crucial role in a variety of cancers. This study was designed to investigate the function and molecular mechanism of lncRNA TTN-AS1 in NSCLC.
Methods: The expression levels of TTN-AS1, miR-491-5p and zinc finger protein 503 (ZNF503) were examined by quantitative real-time polymerase chain reaction (qRT-PCR) or Western blot assay, respectively. Cell viability was detected by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Cell migration and invasion were assessed by transwell assay. Epithelial-to-mesenchymal transition (EMT)-related proteins were measured using Western blot assay. The relationship between TTN-AS1, miR-491-5p and ZNF503 was predicted by starBase2.0 and confirmed by dual-luciferase reporter assay. Xenograft tumor experiment was conducted to analyze the tumor growth in vivo.
Results: The levels of TTN-AS1 and ZNF503 were up-regulated, while miR-491-5p expression was reduced in NSCLC tissues and cells. Knockdown of TTN-AS1 or ZNF503 suppressed cell proliferation, migration, invasion and EMT in NSCLC cells. Overexpression of ZNF503 reversed the effect of TTN-AS1 silencing on NSCLC progression. TTN-AS1 could modulate the expression of ZNF503 via sponging miR-491-5p. Furthermore, TTN-AS1 induced tumor growth in vivo.
Conclusion: Inhibition of TTN-AS1 hindered cell proliferation, migration, invasion and EMT in NSCLC cells by modulating miR-491-5p/ZNF503 axis, providing a promising biomarker for NSCLC treatment.
Keywords: TTN-AS1, miR-491-5p, ZNF503, progression, non-small cell lung cancer
Introduction
Lung cancer is one of the most prevalent malignant tumors, making an essential contribution to cancer mortality.1 Non-small cell lung cancer (NSCLC) accounts for 85% of all lung cancers.2 The overall prognosis of NSCLC remains very poor, with a five-year survival rate below 16%.3 Hence, it is imperative to dig deep into the molecular mechanism of NSCLC to guide clinical diagnosis and treatment to improve the poor prognosis of NSCLC.
Long non-coding RNAs (lncRNAs) are a type of transcripts with more than 200 nucleotides.4 LncRNAs are abnormally expressed in many cancers and participate in tumor initiation and development.5 For example, lncRNA CASC11 facilitated the progression of lung cancer via sponging microRNA-302 and elevating CDK1 expression.6 LncRNA HOXA-AS2 induced the development of NSCLC by targeting microRNA-216a-5p.7 Furthermore, lncRNA TTN antisense RNA1 (TTN-AS1) has been identified as an oncogene. For instance, lncRNA TTN-AS1 contributed to cervical cancer growth and metastasis by modulating miR-573/E2F3 axis.8 In papillary thyroid cancer, lncRNA TTN-AS1 expedited tumor progression via binding with miR-153-3p to up-regulate ZNRF2.9 Additionally, lncRNA TTN-AS1 triggered lung adenocarcinoma cell metastasis via binding to miR-4677-3p and enhancing ZEB1 expression.10 Nevertheless, the role of TTN-AS1 in NSCLC remains to be further studied.
MicroRNAs (miRNAs) are a kind of short non-coding RNAs with 21–25 nucleotides in length.11 Accumulating evidence revealed that miRNAs occupy a crucial position in many biological processes.12 For example, miR-199a-5p hindered cell proliferation and cell cycle via targeting MAP3K11 in NSCLC.13 Kang et al suggested that miR-612 repressed NSCLC development via down-regulating BRD4 expression.14 Moreover, emerging evidence validated that miR-491-5p was a tumor-suppressing factor in various tumors, such as colorectal cancer, osteosarcoma, and gastric cancer.15–17 In lung cancer, miR-491-5p impeded the progression of NSCLC via targeting IGF2BP1.18 However, the relationship between TTN-AS1 and miR-491-5p remains unclear.
Zinc finger protein 503 (ZNF503) contributes to tumorigenesis. A previous research revealed that ZNF503 promoted the progression of hepatocellular carcinoma by repressing GATA3 and being regulated by microRNA-495.19 Nevertheless, the relationship between ZNF503 and miR-491-5p has not been reported.
In the current study, we detected the expression of TTN-AS1 in NSCLC tissues and cells and explored the functional effect of TTN-AS1 on NSCLC development. Furtherly, we researched the molecular mechanism of TTN-AS1 in NSCLC.
Patients and Methods
Sample Collection
Fifty NSCLC samples and adjacent normal tissues were obtained from Huaihe Hospital, Henan University. This study was authorized by the Ethics Committee of Huaihe Hospital, Henan University. All participants were informed of the purpose of this study and signed written informed consent. The clinical characteristics of NSCLC patients are summarized in Table 1.
 |
Table 1 Correlation Between TTN-AS1 Expression and Clinical Characteristics in NSCLC Patients (n=50) |
Cell Culture
NSCLC cell lines (H460, H1299, and A549) and human lung epithelial cell line BEAS-2B were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The PC9 cell line was purchased from BinSuiBio (Shanghai, China). Cells were incubated at 37°C in Dulbecco’s Modified Eagle Medium (DMEM; Solarbio, Shanghai, China) supplemented with 10% fetal bovine serum (FBS; Solarbio).
Cell Transfection
Small interfering RNA (siRNA) targeting TTN-AS1 (si-TTN-AS1), siRNA against ZNF503 (si-ZNF503), siRNA negative control (si-con), TTN-AS1 overexpression vector (pcDNA-TTN-AS1), ZNF503 overexpression vector (pcDNA-ZNF503), the empty overexpression vector (pcDNA), miR-491-5p mimic (miR-491-5p) and the mimic control (miR-con) were synthesized from Ribobio (Guangzhou, China). When cell confluence reached 70%, the vectors and oligonucleotides were transfected into NSCLC cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Lentivirus Infection
Lentivirus vectors containing short hairpin RNA (shRNA) against TTN-AS1 (sh-TTN-AS1) or negative control (sh-con) were constructed by GenePharma (Shanghai, China). When cell confluence reached 70%, 1×106 TU/mL lentivirus supplemented with polybrene were infected into A549 cells. Next, puromycin was used to select stable cell clones.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using Trizol (Invitrogen). The cDNA was synthesized by FastQuant RT Kit (Tiangen, Beijing, China) or miScript Reverse Transcription Kit (Qiagen, Frankfurt, Germany). Then, SYBR Green PCR Master Mix (LMAI Bio, Shanghai, China) was used to perform quantitative PCR. The expression of TTN-AS1 and ZNF503 was normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and miR-491-5p expression was normalized by U6. The primers were as follows: TTN-AS1-F: 5ʹ-CGGGAACAAGCCCTGTG-3ʹ, TTN-AS1-R, 5ʹ-CCGGCCCAAAGATGATG-3ʹ; miR-491-5p-F: 5ʹ-GGAGTGGGGAACCCTTCC-3ʹ, miR-491-5p-R, 5ʹ-GTGCAGGGTCCGAGGT-3ʹ; ZNF503-F: 5ʹ-CAAACTCTCCTCGGTTGCCT-3ʹ, ZNF503-R, 5ʹ-GGGTTTGGAGTACGGCTTGA-3ʹ; GAPDH-F: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ, GAPDH-R, 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ; U6-F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, U6-R, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Western Blot Assay
After extracting the proteins using RIPA buffer (Solarbio), the protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Then, the membrane was incubated with primary antibodies (1:1000; Abcam, Cambridge, UK), followed by incubation with goat anti-rabbit secondary antibody (ab97080, 1:4000; Abcam) for 2 h at room temperature. Finally, the signal intensity was detected by enhanced chemiluminescence reagents (Millipore). The primary antibodies included ZNF503 (ab254715, Abcam), E-cadherin (ab15148, Abcam), N-cadherin (ab18203, Abcam), Vimentin (ab137321, Abcam) and GAPDH (ab9385, Abcam).
Cell Viability
Cells (2.0×103) were injected into 96-well plates. Then, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) solution (Solarbio) was added to each well after incubation for 0 h, 24 h, 48 h, and 72 h. After incubation for another 4 h, dimethyl sulfoxide (DMSO; Solarbio) was added to dissolve formazan crystal. Cell viability was assessed by monitoring the absorbance at 490 nm using a Microplate Reader (Bio-Rad, Hercules, CA, USA).
Transwell Assay
For cell migration assay, cells were placed in the upper chamber with serum-free medium. Besides, the lower chamber was added with 10% FBS (Solarbio). After 24 h of incubation, the migrated cells were treated with methanol and stained with crystal violet for 20 min. For cell invasion assay, transwell chambers were coated with Matrigel (BD Biosciences, San Diego, CA, USA), and other method steps were followed by cell migration assay.
Dual-Luciferase Reporter Assay
The sequences of TTN-AS1 or ZNF503 3ʹUTR containing wild-type or mutant binding sites of miR-491-5p were inserted into pmirGLO vector (Promega, Madison, WI, USA) to construct WT-TTN-AS1, MUT-TTN-AS1, WT-ZNF503 or MUT-ZNF503, respectively. Then, the corresponding luciferase reporter and miR-491-5p mimic or miR-con were cotransfected into NSCLC cells. Finally, Dual-Lucy Assay Kit (Solarbio) was utilized to evaluate the luciferase activity.
Xenograft Tumor Experiment
The BALB/c nude mice used to construct xenograft models were divided into two groups (n=6 per group). A549 cells were infected with lentivirus harboring sh-con or sh-TTN-AS1, respectively. Then, A549 cells were subcutaneously injected into the left of nude mice (5-week-old). Tumor volume was measured every 7 days. Four weeks later, the xenografts were removed, photographed and weighed. The levels of TTN-AS1, miR-491-5p, and ZNF503 were detected by qRT-PCR. The protein level of ZNF503 was measured using Western blot assay. The xenograft assay was ratified by the Animal Welfare Committee of Huaihe Hospital, Henan University. Animal experiments complied with the ARRIVE guidelines and followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Statistical Analysis
GraphPad Prism 7 software (GraphPad Inc., La Jolla, CA, USA) was executed to analyze the data. Data were expressed as mean ± standard deviation. Differences were assessed using Student’s t-test or one-way analysis of variance (ANOVA). Spearman correlation coefficient was conducted to analyze the linear relationship. P<0.05 was considered statistically significant.
Results
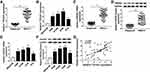
TTN-AS1 and ZNF503 Were Up-Regulated in NSCLC Tissues and Cells
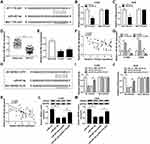
First, we examined the levels of TTN-AS1 and ZNF503 in NSCLC tissues and cells. The results suggested that TTN-AS1 expression in NSCLC tissues was strikingly increased in comparison with adjacent normal tissues (Figure 1A). Similarly, TTN-AS1 expression in NSCLC cells (H460, H1299, A549, and PC9) was drastically raised compared to human lung epithelial cell line BEAS-2B (Figure 1B). In addition, we analyzed the correlation between TTN-AS1 expression and clinical characteristics. The results showed that TTN-AS1 expression was not associated with age, but was related to tumor size, TNM stage and lymph node metastasis (Table 1). Furthermore, the mRNA and protein levels of ZNF503 in NSCLC tissues were substantially higher than that in adjacent normal tissues (Figure 1C and D). Additionally, the mRNA and protein levels of ZNF503 in NSCLC cell lines were evidently elevated compared with BEAS-2B cells (Figure 1E and F). TTN-AS1 expression was positively correlated with ZNF503 expression in NSCLC tissues (Figure 1G). All these data reflected that TTN-AS1 and ZNF503 might play a crucial role in NSCLC development.
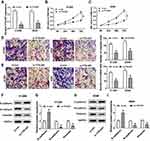
Depletion of TTN-AS1 Impeded Proliferation, Migration, Invasion, and EMT of NSCLC Cells
To investigate the functional role of TTN-AS1 in NSCLC, TTN-AS1 expression was inhibited by transfection with si-TTN-AS1. The results of qRT-PCR exhibited that TTN-AS1 expression was remarkably reduced in the si-TTN-AS1 group compared to the control group (Figure 2A). MTT assay revealed that suppression of TTN-AS1 distinctly restrained the viability of NSCLC cells (Figure 2B and C). Transwell assay demonstrated that cell migration and invasion were prominently limited by knockdown of TTN-AS1 (Figure 2D and E). Moreover, Western blot analysis was carried out to detect the expression of EMT-related proteins, and the results showed that inhibition of TTN-AS1 resulted in an obvious increase in the epithelial marker E-cadherin and a distinct decrease in the mesenchymal markers (N-cadherin and Vimentin) in NSCLC cells (Figure 2F-I). These data demonstrated that knockdown of TTN-AS1 inhibited cell proliferation, migration, invasion and EMT in NSCLC cells.
Silencing of ZNF503 Hindered Proliferation, Migration, Invasion, and EMT of NSCLC Cells
To explore the role of ZNF503 in NSCLC, H1299 and A549 cells were transfected with si-con or si-ZNF503, respectively. The results of qRT-PCR and Western blot revealed that ZNF503 silencing effectively restrained ZNF503 expression in NSCLC cells (Figure 3A and B). MTT assay indicated that cell viability was remarkably reduced in the si-ZNF503 group compared to the si-con group (Figure 3C and D). Transwell assay exhibited that cell migration and invasion were conspicuously suppressed in NSCLC cells transfected with si-ZNF503 compared to the si-con group (Figure 3E and F). Additionally, the level of E-cadherin was markedly increased, and the levels of N-cadherin and Vimentin were especially decreased in the si-ZNF503 group compared with the si-con group (Figure 3G-J). These data demonstrated that ZNF503 knockdown restrained cell proliferation, migration, invasion and EMT in NSCLC cells.
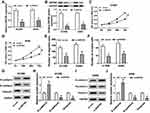
Overexpression of ZNF503 Abrogated the Inhibitory Effect of TTN-AS1 Depletion on NSCLC Progression
Firstly, inhibition of TTN-AS1 dramatically reduced the protein level of ZNF503, and overexpression of TTN-AS1 significantly enhanced the protein level of ZNF503 (Figure 4A and B). We also found that the protein expression of ZNF503 was strikingly elevated in NSCLC cells transfected with pcDNA-ZNF503 compared to the pcDNA group (Figure 4C). In addition, to investigate the role of TTN-AS1 and ZNF503 in NSCLC progression, H1299 and A549 cells were transfected with si-con, si-TTN-AS1, si-TTN-AS1+pcDNA or si-TTN-AS1+pcDNA-ZNF503, respectively. MTT assay suggested that knockdown of TTN-AS1 substantially impeded cell viability, while this effect was reversed by up-regulating ZNF503 expression (Figure 4D and E). Transwell assay confirmed that depletion of TTN-AS1 evidently hindered cell migration and invasion, whereas this effect was restored after transfection with pcDNA-ZNF503 (Figure 4F and G). Furthermore, TTN-AS1 silencing led to a distinct increase in the expression of E-cadherin and a marked decrease in the levels of N-cadherin and Vimentin, which were reversed by transfection with ZNF503 overexpression (Figure 4H-K). Taken together, these data demonstrated that up-regulation of ZNF503 could abrogate the inhibitory effect of TTN-AS1 knockdown on NSCLC progression.
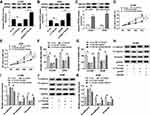
TTN-AS1 Targeted miR-491-5p to Regulate the Expression of ZNF503
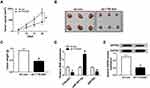
In order to explore the underlying molecular mechanisms of TTN-AS1 in NSCLC, the starBase v2.0 online database predicted that TTN-AS1 and miR-491-5p had putative binding sites (Figure 5A). Besides, dual-luciferase reporter assay was applied to verify the interaction between TTN-AS1 and miR-491-5p. The results confirmed that cotransfection of miR-491-5p mimic and WT-TTN-AS1 prominently reduced the luciferase activity in H1299 and A549 cells (Figure 5B and C). The expression of miR-491-5p was observably reduced in NSCLC tissues and cells (Figure 5D and E). Spearman correlation analysis manifested that TTN-AS1 expression was negatively associated with miR-491-5p expression (Figure 5F). Subsequently, TTN-AS1 silencing observably facilitated miR-491-5p expression, and TTN-AS1 overexpression predominantly restrained miR-491-5p expression (Figure 5G). Furtherly, we also analyzed the relationship between miR-491-5p and ZNF503. The starBase2.0 online database predicted that miR-491-5p and ZNF503 3ʹUTR had putative binding sites (Figure 5H). Dual-luciferase reporter assay validated that miR-491-5p mimic dramatically decreased the luciferase activity of WT-ZNF503, which was restored after transfection with pcDNA-TTN-AS1 (Figure 5I and J). Moreover, the expression of miR-491-5p and ZNF503 was negatively correlated in NSCLC tissues (Figure 5K). Additionally, the protein level of ZNF503 was obviously reduced in the miR-491-5p group compared with the miR-con group, whereas this effect was abolished by up-regulating TTN-AS1 expression (Figure 5L and M). These data disclosed that TTN-AS1 modulated ZNF503 expression via sponging miR-491-5p in NSCLC cells.
Knockdown of TTN-AS1 Blocked Tumor Growth in vivo
To evaluate the effect of TTN-AS1 on NSCLC tumorigenesis in vivo, we established a xenograft model. Firstly, A549 cells transfected with sh-con or sh-TTN-AS1 were subcutaneously injected into the left of nude mice. Compared with the negative control group, tumor volume and weight in the sh-TTN-AS1 group were prominently decreased (Figure 6A-C). The levels of TTN-AS1, miR-491-5p and ZNF503 were examined by qRT-PCR. The results indicated that the levels of TTN-AS1 and ZNF503 were remarkably reduced, while the expression of miR-491-5p was drastically increased in the sh-TTN-AS1 group compared to the negative control group (Figure 6D). Similarly, the protein level of ZNF503 was overtly decreased in the sh-TTN-AS1 group compared with the sh-con group (Figure 6E). Thus, these data concluded that TTN-AS1 knockdown suppressed tumor growth in vivo.
Discussion
Emerging evidence has demonstrated that lncRNAs mediate tumor invasion and metastasis to act as promising biomarkers in tumor therapy.20 For example, Qu et al revealed that lncRNA CASC19 facilitated the development of NSCLC by sponging miR-130b-3p and increasing ZEB2 expression.21 Ge et al indicated that lncRNA ZFAS1 contributed to NSCLC growth by binding to microRNA-193a-3p.22 Previous investigations suggested that the expression of TTN-AS1 was overtly elevated in many cancers, including lung cancer.10 Dong et al found that lncRNA TTN-AS1 facilitated gastric cancer tumorigenesis through sponging miR-376b-3p.23 Moreover, lncRNA TTN-AS1 induced lung adenocarcinoma progression by regulating miR-142-5p/cyclin-dependent kinase 5 (CDK5) axis.24 What’s more, lncRNA TTN-AS1 contributed to the development of lung adenocarcinoma via inhibiting the level of PTEN protein and activating the PI3K/AKT pathway.25 Previous studies have clarified the role of TTN-AS1 in lung adenocarcinoma. However, this study revealed that TTN-AS1 was highly expressed and promoted tumor progression by regulating cell proliferation, migration, invasion and EMT in NSCLC.
LncRNAs could function as competing endogenous RNAs to repress the expression of target genes.26 Additionally, it has been reported that miRNAs bound to the 3ʹUTR of the target genes, thereby impeding mRNA expression.27 Hence, the online database was used to predict the targets of TTN-AS1. In previous studies, miR-491-5p was a tumor inhibitor and undermined tumor growth and progression by down-regulating JMJD2B in gastric cancer.28 MiR-491-5p restricted tumorigenesis and growth via directly binding to Notch3 3ʹUTR in nasopharyngeal carcinoma.29 In cervical cancer, miR-491-5p repressed tumor development via enhancing hTERT and modulating the PI3K/AKT pathway.30 More importantly, miR-491-5p blocked cell proliferation and invasion via targeting IGF2BP1 in NSCLC.18 According to the existing researches, miR-491-5p was selected as the research target. Consistent with previous studies, miR-491-5p was down-regulated in NSCLC. Furthermore, our research first confirmed the targeting relationship between TTN-AS1 and miR-491-5p, and speculated that TTN-AS1 modulated NSCLC development via sponging miR-491-5p.
ZNF503 is an evolutionarily conserved zinc finger protein.31 Recent evidence suggested that miR-340-5p hindered non-small cell lung cancer progression through degradation of ZNF503.32 In the current study, ZNF503 was distinctly up-regulated in NSCLC. More importantly, ZNF503 overexpression reversed the inhibitory effect of TTN-AS1 depletion on NSCLC progression. Besides, TTN-AS1 regulated ZNF503 expression via sponging miR-491-5p in NSCLC cells.
Conclusion
In conclusion, TTN-AS1 facilitated NSCLC progression via sponging miR-491-5p and elevating ZNF503 expression. Therefore, lncRNA TTN-AS1 might be a potential therapeutic target for lung cancer treatment.
Highlights
- TTN-AS1 and ZNF503 were up-regulated in NSCLC tissues and cells.
- Knockdown of TTN-AS1 or ZNF503 inhibited NSCLC progression.
- TTN-AS1 promoted NSCLC progression by regulating ZNF503.
- TTN-AS1 targeted miR-491-5p to elevate ZNF503 expression.
- TTN-AS1 facilitated tumor growth in vivo.
Disclosure
The authors declare that they have no financial conflicts of interest.
References
1. Seijo LM, Peled N, Ajona D, et al. Biomarkers in lung cancer screening: achievements, promises, and challenges. J Thorac Oncol. 2019;14(3):343–357. doi:10.1016/j.jtho.2018.11.023
2. Relli V, Trerotola M, Guerra E, Alberti S. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019;25(7):585–594. doi:10.1016/j.molmed.2019.04.012
3. Balata H, Fong KM, Hendriks LE, et al. Prevention and early detection for NSCLC: advances in thoracic oncology 2018. J Thorac Oncol. 2019;14(9):1513–1527. doi:10.1016/j.jtho.2019.06.011
4. Fan C, Tang Y, Wang J, et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16(1):130. doi:10.1186/s12943-017-0699-3
5. Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28(4):287–301. doi:10.1016/j.tcb.2017.11.008
6. Tong W, Han TC, Wang W, Zhao J. LncRNA CASC11 promotes the development of lung cancer through targeting microRNA-302/CDK1 axis. Eur Rev Med Pharmacol Sci. 2019;23(15):6539–6547. doi:10.26355/eurrev_201908_18539
7. Cui TJ, Lin GS, Dai YM, et al. LncRNA HOXA-AS2 regulates microRNA-216a-5p to promote malignant progression of non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23(3Suppl):264–273. doi:10.26355/eurrev_201908_18656
8. Chen P, Wang R, Yue Q, Hao M. Long non-coding RNA TTN-AS1 promotes cell growth and metastasis in cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun. 2018;503(4):2956–2962. doi:10.1016/j.bbrc.2018.08.077
9. Cui Z, Luo Z, Lin Z, Shi L, Hong Y, Yan C. Long non-coding RNA TTN-AS1 facilitates tumorigenesis of papillary thyroid cancer through modulating the miR-153-3p/ZNRF2 axis. J Gene Med. 2019;21(5):e3083. doi:10.1002/jgm.3083
10. Zhong Y, Wang J, Lv W, Xu J, Mei S, Shan A. LncRNA TTN-AS1 drives invasion and migration of lung adenocarcinoma cells via modulation of miR-4677-3p/ZEB1 axis. J Cell Biochem. 2019;120(10):17131–17141. doi:10.1002/jcb.28973
11. Wu KL, Tsai YM, Lien CT, Kuo PL, Hung AJ. The roles of MicroRNA in lung cancer. Int J Mol Sci. 2019;20:7. doi:10.3390/ijms20071611
12. Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. 2019;11(1):25. doi:10.1186/s13148-018-0587-8
13. Li Y, Wang D, Li X, et al. MiR-199a-5p suppresses non-small cell lung cancer via targeting MAP3K11. J Cancer. 2019;10(11):2472–2479. doi:10.7150/jca.29426
14. Kang X, Kong F, Wu S, et al. microRNA-612 suppresses the malignant development of non-small-cell lung cancer by directly targeting bromodomain-containing protein 4. Onco Targets Ther. 2019;12:4167–4179. doi:10.2147/OTT.S204004
15. Lu L, Cai M, Peng M, Wang F, Zhai X. miR-491-5p functions as a tumor suppressor by targeting IGF2 in colorectal cancer. Cancer Manag Res. 2019;11:1805–1816. doi:10.2147/CMAR.S183085
16. Chen T, Li Y, Cao W, Liu Y. miR-491-5p inhibits osteosarcoma cell proliferation by targeting PKM2. Oncol Lett. 2018;16(5):6472–6478. doi:10.3892/ol.2018.9451
17. Yu T, Wang LN, Li W, et al. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018;109(5):1393–1403. doi:10.1111/cas.13583
18. Gong F, Ren P, Zhang Y, Jiang J, Zhang H. MicroRNAs-491-5p suppresses cell proliferation and invasion by inhibiting IGF2BP1 in non-small cell lung cancer. Am J Transl Res. 2016;8(2):485–495.
19. Yin G, Liu Z, Wang Y, et al. ZNF503 accelerates aggressiveness of hepatocellular carcinoma cells by down-regulation of GATA3 expression and regulated by microRNA-495. Am J Transl Res. 2019;11(6):3426–3437.
20. Jiang C, Li X, Zhao H, Liu H. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer. 2016;15(1):62. doi:10.1186/s12943-016-0545-z
21. Qu CX, Shi XC, Zai LQ, Bi H, Yang Q. LncRNA CASC19 promotes the proliferation, migration and invasion of non-small cell lung carcinoma via regulating miRNA-130b-3p. Eur Rev Med Pharmacol Sci. 2019;23(3 Suppl):247–255. doi:10.26355/eurrev_201908_18654
22. Ge HB, Chen S, Huang SR, Zhu J. Long noncoding RNA ZFAS1 acts as an oncogene by targeting miR-193a-3p in human non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23(15):6516–6523. doi:10.26355/eurrev_201908_18535
23. Dong MM, Peng SJ, Yuan YN, Luo HP. LncRNA TTN-AS1 contributes to gastric cancer progression by acting as a competing endogenous RNA of miR-376b-3p. Neoplasma. 2019;66(4):564–575. doi:10.4149/neo_2018_180927N721
24. Jia Y, Duan Y, Liu T, et al. LncRNA TTN-AS1 promotes migration, invasion, and epithelial mesenchymal transition of lung adenocarcinoma via sponging miR-142-5p to regulate CDK5. Cell Death Dis. 2019;10(8):573. doi:10.1038/s41419-019-1811-y
25. Luo J, Liu Z. Long non-coding RNA TTN-AS1 promotes the progression of lung adenocarcinoma by regulating PTEN/PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2019;514(1):140–147. doi:10.1016/j.bbrc.2019.04.050
26. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):5. doi:10.3390/ijms19051310
27. Takahashi RU, Prieto-Vila M, Kohama I, Ochiya T. Development of miRNA-based therapeutic approaches for cancer patients. Cancer Sci. 2019;110(4):1140–1147. doi:10.1111/cas.13965
28. Zhang J, Ren J, Hao S, et al. MiRNA-491-5p inhibits cell proliferation, invasion and migration via targeting JMJD2B and serves as a potential biomarker in gastric cancer. Am J Transl Res. 2018;10(2):525–534.
29. Zhang Q, Li Q, Xu T, Jiang H, Xu LG. miR-491-5p suppresses cell growth and invasion by targeting Notch3 in nasopharyngeal carcinoma. Oncol Rep. 2016;35(6):3541–3547. doi:10.3892/or.2016.4713
30. Zhao Q, Zhai YX, Liu HQ, Shi YA, Li XB. MicroRNA-491-5p suppresses cervical cancer cell growth by targeting hTERT. Oncol Rep. 2015;34(2):979–986. doi:10.3892/or.2015.4013
31. Shahi P, Slorach EM, Wang CY, et al. The transcriptional repressor ZNF503/Zeppo2 promotes mammary epithelial cell proliferation and enhances cell invasion. J Biol Chem. 2015;290(6):3803–3813. doi:10.1074/jbc.M114.611202
32. Lu G, Zhang Y. MicroRNA-340-5p suppresses non-small cell lung cancer cell growth and metastasis by targeting ZNF503. Cell Mol Biol Lett. 2019;24(1):34. doi:10.1186/s11658-019-0161-1
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.