Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA TBX5-AS1 Regulates the Tumor Progression Through the PI3K/AKT Pathway in Non-Small Cell Lung Cancer
Received 27 March 2020
Accepted for publication 10 July 2020
Published 12 August 2020 Volume 2020:13 Pages 7949—7961
DOI https://doi.org/10.2147/OTT.S255195
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Qing-hai Qu,1,* Shui-zheng Jiang,2,* Xin-ying Li3
1Department of Blood Transfusion, Weifang Yidu Center Hospital, Weifang Medical University, Qingzhou, Shandong 262500, People’s Republic of China; 2Calling Ethos Construction Transfusion, Weifang Yidu Center Hospital, Weifang Medical University, Qingzhou, Shandong 262500, People’s Republic of China; 3Department of Conservative Dentistry and Endodontics, Weifang Dental Hospital, Qingzhou, Shandong 262500, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qing-hai Qu Email [email protected]
Purpose: Long non-coding RNAs (lncRNAs) have been reported to play important roles in tumor biology. In this study, we aimed to investigate the effects of T-box transcription factor 5 antisense RNA 1 (TBX5-AS1) on aggressive phenotypes of non-small cell lung cancer (NSCLC) cells and explore its regulatory pathway.
Methods: The expression of TBX5-AS1 in tissues, plasma, and cells was determined by qRT-PCR. Cell viability, proliferation, migration, invasion, and apoptosis were assessed using MTT, colony formation, wound-healing, Transwell, and flow cytometry assay, respectively. Western blot analysis was performed to measure the expression of apoptosis-related proteins. Besides, transfected cells were exposed to PI3K activator (740Y-P) to verify the regulatory pathway.
Results: TBX5-AS1 expression was down-regulated in NSCLC tissues, plasma, and cells, and associated with lymph node metastasis and histological grade. Overexpression of TBX5-AS1 inhibited cell viability, colony formation, migration, and invasion, while it promoted apoptosis. Conversely, knockdown of TBX5-AS1 showed the completely opposite results. Additionally, western blot showed that the phosphorylation of PI3K and AKT was stimulated by TBX5-AS1 knockdown and suppressed by TBX5-AS1 overexpression. The addition of 740Y-P in transfected cells reversed the TBX5-AS1-induced inhibition of PI3K and AKT phosphorylation and effects on aggressive phenotypes of NSCLC cells.
Conclusion: The study confirmed the down-regulation of TBX5-AS1 in patients with NSCLC and its association with the progression. We innovatively proposed a possible model of TBX5-AS1-mediated gene regulation in NSCLC progression that TBX5-AS1 inhibited the aggressive phenotypes of NSCLC cells through inactivating the PI3K/AKT pathway. This finding provided a novel insight into NSCLC pathogenesis.
Keywords: non-small cell lung cancer, long non-coding RNAs, T-box transcription factor 5 antisense RNA 1, aggressive phenotype, PI3K/AKT pathway
Introduction
Non-small cell lung cancer (NSCLC) as the most common type of lung cancer has been the leading cause of cancer-related death worldwide.1,2 Currently, various therapeutic strategies including radiation, chemotherapy, neoadjuvant chemotherapy, and surgery separately or in combination are widely applied in the therapy of NSCLC.3 Fortunately, many patients obtain unprecedented survival benefits from the clinical use of small molecule tyrosine kinase inhibitors and immunotherapy.4 Despite significant advancements in NSCLC treatment being achieved over the past two decades, the overall cure and 5-year survival rates for NSCLC remain low, especially in metastatic lung cancer.5,6 Thus, understanding of the pathogenic mechanisms of NSCLC progression and finding novel therapeutic targets are required to expand the clinical benefit to a broader patient population.
Long non-coding RNAs (lncRNAs) are increasingly implicated as gene regulators in diverse diseases.7–9 Notably, lncRNAs are considered to play important roles in disease progression of various tumors, such as NSCLC, breast cancer, gastric cancer, bladder cancer, and many other cancer types.10,11 Accordingly, wide-range evidences have demonstrated that lncRNAs are emerging as the potential therapeutic target for cancer patients.12,13 More recently, T-box transcription factor 5 (TBX5) serves as a novel tumor suppressor gene in the disease progression.14 TBX5 antisense RNA 1 (TBX5-AS1), a newly discovered lncRNA, is the tail-to-tail antisense of TBX5.15 It is worth noting that TBX5-AS1 may be associated with the prognosis of patients with lung squamous cell carcinoma (LUSC).16 In addition, Zhang et al also reported the TBX5-AS1 might have an effect on the regulation of lung adenocarcinoma development.17 With these evidences, we supposed that TBX5-AS1 might be involved in the progression of NSCLC and had the potential to be a potent therapeutic target for NSCLC therapy.
The phosphatidylinositol 3-kinase (PI3K)/AKT pathway is recognized as a pivotal signaling pathway in carcinogenesis and regulates various key cellular functions, such as proliferation, survival, and metabolism.18,19 Genes in this pathway are the most frequently amplified, mutated, and translocated in human cancers.20 Research shows that aberrant activation of the PI3K/AKT pathway is associated with tumor genesis, cancer progression, and drug resistance.21 Previously, TBX3 as another member of TBX family was found to be associated with the PI3K/AKT pathway.22 Moreover, it is demonstrated that TBX1 functions as a tumor suppressor in thyroid cancer by inhibiting activation of the PI3K/AKT pathway.23 Therefore, we speculated that TBX5-AS1 might participate in NSCLC progression through the PI3K/AKT pathway. However, no evidence demonstrated the role and regulatory pathway of TBX5-AS1 in NSCLC progression. Thus, in the present study, we detected the aberrant expression of TBX5-AS1 in NSCLC and further verified its effects on aggressive phenotypes of NSCLC cells. More importantly, the regulation effect of TBX5-AS1 on the PI3K/AKT pathway was innovatively studied in vitro.
Materials and Methods
Clinical Samples
Ninety-six pairs of NSCLC tissues and para-carcinoma normal tissues were originally obtained from 96 patients with NSCLC in our hospital, between January 2018 and December 2019. In addition, the plasma samples were respectively collected from 96 NSCLC patients and 96 healthy volunteers. All enrolled patients did not receive surgery, radiotherapy, or chemotherapy before tissue/serum collection. All clinical samples were immediately frozen in liquid nitrogen and stored at −80 °C in a refrigerator until the analysis of TBX5-AS1 expression. All protocols have been approved by the Ethics Committee of Weifang Yidu Center Hospital and performed in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants.
Cell Lines and Cell Culture
The human bronchial epithelioid cell line (16HBE) and human lung cancer cell lines (A549, H1299 and NCI-H520) were originally obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). All cell lines were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Cambridge, MA, USA) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL). Cell lines were incubated in a humidified incubator at 37 °C with a 5% CO2 atmosphere.
RNA Extraction and qRT-PCR
Total RNA from tumor tissues and cell lines was prepared using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Total RNA from plasma was extracted using TRIzol LS Reagent (Invitrogen) according to the manufacturer’s protocol. A NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) was used to determine the purity and concentration of RNA, and agarose gel electrophoresis was used to assess the integrity of RNA. The samples with absorbance ratios 260/280 (1.8–2.0) and 260/230 (1.9–2.2) were considered for inclusion in the study. Subsequently, the reverse transcription of RNA to cDNA was performed using Reverse Transcription System (Promega Corporation, Madison, WI, USA) following the manufacturer’s protocol. Finally, the qRT-PCR assays were carried out on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the SYBR PrimeScript RT-PCR Kit (Takara Bio, Dalian, China). The relative expression level was analyzed using the comparative cycle threshold (CT) (2−ΔΔCT) method, using β-actin as the endogenous control. PCR primer sequences were listed as follows: 1) TBX5-AS1 forward 5ʹ-GCAAAACGTACCGCGTAGAC-3ʹ and reverse 5ʹ-GATGCCGGAGAAAGGAACCA-3ʹ; and 2) β-actin forward 5ʹ- CACCATTGGCAATGAGCGGTTC-3ʹ and reverse 5ʹ-AGGTCTTTGCGGATGTCCACGT-3ʹ.
Cell Transfection
All transfections were performed using the Lipofectamine 2000 Transfection Kit (Invitrogen) according to the manufacturer’s instruction. The knockdown of TBX5-AS1 in NCI-H520 cells was induced by small interfering RNA (siRNA) targeting TBX5-AS1 (si-TBX5-AS1), using scramble siRNA as a negative control (si-NC). Besides, human TBX5-AS1 cDNA insert was cloned into the pcDNA3.1 plasmid (Invitrogen) to construct a recombinant plasmid vector with the TBX5-AS1 gene (pcDNA3.1-TBX5-AS1). Then, pcDNA3.1-TBX5-AS1 was used to induce the overexpression of TBX5-AS1 in A549 cells, using pcDNA3.1 plasmid as a negative control (pcDNA3.1-NC). After transfection, the aberrant expression of TBX5-AS1 was verified by qRT-PCR.
MTT Assay
Cell proliferation was evaluated by MTT assay using the Cell Proliferation Reagent Kit I (Roche, Basel, Switzerland) according to the manufacturer’s protocol. Briefly, the prepared cells (1×104 cells/well) were seeded into 96-well plates and incubated for 0, 1, 2, 3, 4 d at 37 °C with a 5% CO2 atmosphere. After the incubation, 10 μL MTT solution was added into each well, followed by incubation for another 4 h. Then, the medium was removed and 100 μL solubilization solution was added into each well and incubated overnight. Finally, the absorbance value was detected at 450 nm using a microplate reader (BioRad, Hercules, CA, USA).
Colony Formation Assay
Colony formation ability was evaluated using the standard two-layer soft agar culture.24 In brief, the prepared cells were seeded in the 24-well culture plates at a density of 200 cells/well. Then, cells were maintained in RPMI-1640 medium with 0.6% agarose underlay. After incubation for 14 days, the surviving colonies were fixed with 3.7% acetic acid methanol and then stained with 0.1% crystal violet. Finally, the stained colonies were manually counted and visualized under a microscope (Olympus, Tokyo, Japan).
Cell Migration and Invasion Assay
Cell migration ability was evaluated using a wound-healing assay. The prepared cells were seeded on a 6-well plate in RPMI-1640 medium supplemented with 10% FBS and cultured to at least 90% confluence. Then, cells were starved in serum-free RPMI-1640 and cultured for another 24 h. Subsequently, the monolayer cells were scraped with a 10 µL plastic pipette tip in the middle of a culture plate to make a “Scratch” wound and washed with serum-free medium. At 0 and 48 h after scratching, images of the wounded area were collected by an inverted phase contrast microscope (magnification: 10× objective; Olympus, Tokyo, Japan). The rate of wound area was calculated relative to the initial wound area. After pretreating confluent cell monolayers (si1-TBX5-AS1 and pcDNA3.1-TBX5-AS1) with mitomycin C (MMC, 5 μM; Wako Pure Chemicals Ltd., Osaka, Japan) for 2 h, wound-healing assays were also performed.
Cell invasion ability was evaluated using a Matrigel-coated Transwell chamber (BD Biosciences, San Jose, CA, USA). Briefly, the prepared cells were pre-starved in serum-free medium for 24 h and then incubated in the upper chamber of a Matrigel-coated Transwell (8 μm) for 48 h. RPMI-1640 containing 10% FBS was added into the lower chamber of Transwell. After incubation for 24 h, non-invading cells were removed from the upper surface of the Matrigel. Finally, the lower side of the upper chamber was fixed with cold methanol for 30 min and then stained with 4,6-diamidino-2-phenylindole (Sigma Aldrich, St. Louis, MO, USA). Images were photographed under a microscope (magnification: 400× objective; Olympus, Tokyo, Japan).
Flow Cytometry Assay
Cell apoptosis was evaluated by flow cytometry assay using the Annexin V-fluorescein isothiocyanate (FITC) apoptosis kit (BD Biosciences, San Jose, CA, USA). The prepared cells were double-labeled with Annexin V-FITC (5 μL) and propidium iodide (5 μL) for 15 min following the manufacturer’s protocol. The apoptosis rate was immediately analyzed in a FACSAria™ Fusion flow cytometer using CellQuest software (BD Biosciences, San Jose, CA, USA).
Western Blot
Whole-cell lysates were lysed from cell lines using RIPA buffer (Thermo Scientific, USA). The concentration of lysates was quantified using the Bicinchoninic Acid protein assay kit (Thermo Fisher Scientific, Cambridge, MA, USA). An equal amount of proteins (50 μg) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto the polyvinylidene fluoride (PVDF) membranes (Millipore, USA). After blocking with 5% (w/v) skim milk, the blotting PVDF membranes were separately incubated with specific primary antibodies against Bcl-2, 4223; Bax, 14,796; p-PI3K, 17,366; (1:1000, Cell Signaling Technology, USA), PI3K, ab32089; AKT, ab8805; GAPDH, ab181606 (1/1000, Abcam, USA), and p-AKT, SAB4301414 (1:1000, Sigma-Aldrich, USA) overnight at 4 °C. Subsequently, PVDF membranes were blocked with goat anti-rabbit IgG HRP-linked secondary antibody (1:5000, Abcam) at room temperature for 1 h. The signal bands were measured with an Enhanced Chemiluminescence Kit (Thermo Fisher Scientific, Cambridge, MA, USA) and the band intensity was analyzed by Quantity One software (BioRad, Hercules, CA, USA).
Statistical Analysis
Statistical analysis was analyzed by GraphPad Prism 7 software and SPSS software version 22.0 (IBM Corporation, Armonk, NY, USA). For in vitro experiments, all experiments were repeated in triplicate and the results were expressed as the mean±standard deviation (SD). Student’s t-test or one-way ANOVA followed by Tukey’s multiple comparisons test was performed to analyze the statistical differences between two or among multiple groups. The association between expression level of TBX5-AS1 and clinicopathological parameters of patients were analyzed using the chi-square test. Distinguishing performance of the TBX5-AS1 was assessed by constructing a receiver operating characteristic (ROC) curve and evaluated by calculating the area under curve (AUC). p<0.05 was considered statistically significant.
Results
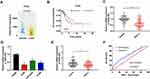
TBX5-AS1 Expression is Down-Regulated in NSCLC and Associated with Progression of NSCLC
Lung adenocarcinoma (LUAD) is one of the common types of NSCLC. In the light of the data from TCGA, we found that TBX5-AS1 expression in LUAD tissues was lower than that of normal tissues (p<0.01, Figure 1A). Moreover, the Kaplan–Meier survival analysis showed that the overall survival in patients with low TBX5-AS1 expression was poorer than that of patients with high TBX5-AS1 expression (p=0.02, Figure 1B). These data indicated that TBX5-AS1 might be correlated with prognosis in NSCLC. To determine the involvement of TBX5-AS1 in the progression of NSCLC, we firstly detected the expression of TBX5-AS1 in tissues, cell lines, and plasma, respectively. As shown in Figure 1C and E (Supplementary Table-1) (Supplementary Table-2), the expression level of TBX5-AS1 both in tissues and plasma from NSCLC patients were reduced compared to the normal and control (p<0.01). Similarly, a lower expression of TBX5-AS1 was observed in NSCLC cell lines (A549, NCI-H520, and H1299) compared to 16HBE cells (p<0.01, Figure 1D). Furthermore, to evaluate the clinical significance of TBX5-AS1 in NSCLC, we assessed its diagnostic performance and the association with pathological parameters. ROC analysis demonstrated that the AUC for the TBX5-AS1 level in discrimination between NSCLC and healthy volunteers was 0.6498 (p=0.0003, Figure 1F), indicating a favourable distinguishing performance of TBX5-AS1 for NSCLC. The chi-square test showed that the TBX5-AS1 expression level was significantly associated with lymph node metastasis (p=0.002) and histological grade (p=0.004), but not associated with gender, age, and tumor size (Table 1). Taken together, the results suggested that the down-regulation of TBX5-AS1 might play an important role in the progression of NSCLC.
 |
Table 1 Association Between TBX5-AS1 Expression and Clinicopathological Parameters of NSCLC Patients |
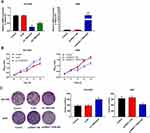
TBX5-AS1 Inhibits the Proliferation of NSCLC Cells
To confirm the role of TBX5-AS1 in the progression of NSCLC, TBX5-AS1 was knocked down or overexpressed in two NSCLC cells, respectively. The expression of TBX5-AS1 was confirmed by qRT-PCR after transfection. As shown in Figure 2A, the expression of TBX5-AS1 was successfully reduced in NCI-H520 cells (p<0.01) and up-regulated in A549 cells (p<0.01). Then, the MTT assay was performed to evaluate the effects of TBX5-AS1 on cell viability. The results showed that the cell viability was significantly enhanced by the inhibition of TBX5-AS1, while decreased by the overexpression of TBX5-AS1 (p<0.01, Figure 2B). Furthermore, we also measured the colony formation ability of NSCLC cells after transfection. Figure 2C reveals similar results with the MTT assay, the colony formation ability of NSCLC cells was promoted by the inhibition of TBX5-AS1 (p<0.01) while suppressed by the overexpression of TBX5-AS1 (p<0.01). Overall, these results indicated that TBX5-AS1 significantly inhibited the cell proliferation in NSCLC cell lines.
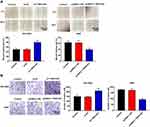
TBX5-AS1 Inhibits the Migration and Invasion of NSCLC Cells
To further verify the effects of TBX5-AS1 on the aggressive phenotype of NSCLC cells, the wound-healing assay and Transwell assay were respectively used to assess the cell migration and invasion ability. We found that cell migration ability (wound healing rate) of NSCLC cells was significantly enhanced by the down-regulation of TBX5-AS1 (p<0.01), while suppressed by the up-regulation of TBX5-AS1 (p<0.01, Figure 3A). To assess whether wound healing was due to cell migration instead of cell proliferation, we detected the wound healing in the transfected group (si1-TBX5-AS1 and pcDNA3.1-TBX5-AS1) and the transfection group with mitomycin C (MMC, an inhibitor of proliferation). Our data showed the wound healing rate in the transfected group was slightly higher than that in the transfection group with MMC (si1-TBX5-AS1+MMC and pcDNA3.1-TBX5-AS1+MMC), but there was no significant difference (Figure S1). Therefore, TBX5-AS1 regulated wound healing by cell migration regardless of proliferation. Moreover, we also found that cell invasion ability of NSCLC cells was enhanced by the down-regulation of TBX5-AS1 (p<0.01), while suppressed by the up-regulation of TBX5-AS1 (p<0.01, Figure 3B). Overall, these results indicated that TBX5-AS1 significantly inhibited the migration and invasion of NSCLC cell lines in vitro.
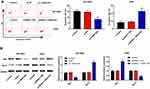
TBX5-AS1 Induces Apoptosis of NSCLC Cells
We also assessed the effects of TBX5-AS1 on cell apoptosis using flow cytometry assay. As shown in Figure 4A, the apoptosis rate of NSCLC cells was significantly decreased by the down-regulation of TBX5-AS1 compared to controls (p<0.01), while increased by the up-regulation of TBX5-AS1 (p<0.01). To further verify the effects of TBX5-AS1 on apoptosis, the expression levels of apoptosis-related proteins Bax and Bcl-2 were detected. Western blot assay demonstrated that the down-regulation of TBX5-AS1 in NSCLC cells significantly inhibited Bax expression (p<0.01) and enhanced Bcl-2 expression (p<0.01, Figure 4B). Conversely, the up-regulation of TBX5-AS1 significantly increased Bax expression (p<0.01) and suppressed Bcl-2 expression (p<0.01, Figure 4B). Overall, these results suggested that TBX5-AS1 significantly induced apoptosis and pro-apoptotic protein expression, while it inhibited the anti-apoptotic protein expression in NSCLC cell lines.
TBX5-AS1 Inhibits the Aggressive Phenotypes of NSCLC Cells Through Inactivating the PI3K/AKT Pathway
To confirm the hypothesis that TBX5-AS1 regulates the PI3K/AKT pathway, we measured the expression levels of PI3K and AKT in transfected NSCLC cells. As shown in Figure 5A, the knockdown of TBX5-AS1 in NCI-H520 cells obviously stimulated the phosphorylation of PI3K and AKT (p<0.01). Inversely, the TBX5-AS1 overexpression in A549 cell suppressed the phosphorylation of PI3K and AKT (p<0.01, Figure 5B).
Subsequently, in order to further confirm the molecular mechanism of TBX5-AS1 in the PI3K/AKT pathway, the PI3K activator (740Y-P) was used to treat the pcDNA3.1-TBX5-AS1 transfected cells. Firstly, western blot assay showed that the addition of 740Y-P in transfected cells reversed the inhibition of PI3K and AKT phosphorylation caused by TBX5-AS1 overexpression (p<0.01, Figure 6A), confirming the 740Y-P-induced activation of the PI3K/AKT pathway. Then, we detected the influences of 740Y-P on the phenotypes of transfected cells, including colony formation, migration, invasion, and apoptosis. The results showed that the addition of 740Y-P significantly reversed the TBX5-AS1-induced inhibition on colony formation, migration, and invasion (p<0.05, Figure 6B–D). Besides, the flow cytometry assay demonstrated that the TBX5-AS1-induced apoptosis was alleviated by 740Y-P treatment (p<0.01, Figure 6E). Overall, these results suggested that TBX5-AS1 inhibited the aggressive phenotypes of NSCLC cells through inactivating the PI3K/AKT pathway.
Discussion
Despite significant developments in the targeted therapies for NSCLC, the prognosis for advanced stage patients remains dismal. Hence, further elaboration of the underlying pathogenic mechanisms for NSCLC progression remains the research challenge.6 In recent years, lncRNAs have been proven to function as tumor suppressors or oncogenes in various tumors.25 Whereas, the lncRNA profiles are extremely complicated and not yet fully elucidated. Thus, the discovery of new lncRNAs will probably change the landscape of cancer genetics and potentially become effective therapeutic targets. Herein, this ground-breaking study identified TBX5-AS1 as a suppressor lncRNA in NSCLC for the first time. We verified the down-regulation of TBX5-AS1 and its association with the progression of NSCLC. More importantly, the present study indicated that TBX5-AS1 inhibited the aggressive phenotypes of NSCLC cells through inactivating the PI3K/AKT pathway.
Numerous studies have proved that lncRNAs play major roles in the gene regulation and thus influence the diverse aspects of cellular homeostasis, including survival, proliferation, or genomic stability.26 The aberrant expression of several lncRNAs is associated with malignant transformation and poor prognosis of cancers.27 TBX5-AS1, a novel cancer-related lncRNA, is the antisense RNA of TBX5. A previous bioinformatics analysis revealed that TBX5-AS1 was significantly down-regulated in female lung cancer tissue.28 Hereby, we detected the differential expression of TBX5-AS1 in clinical samples and cell lines. The results demonstrated that TBX5-AS1 was down-regulated in NSCLC patients and cell lines, which was identified with the previous analysis.28 Furthermore, using a cohort of NSCLC patients, the clinical relationship analysis showed that TBX5-AS1 expression was closely associated with lymph node metastasis and histological grade, suggesting that the down-regulation of TBX5-AS1 led to a poorer clinical outcome. Similarly, a bioinformatics analysis has also reported that TBX5-AS1 may be associated with the prognosis of patients with LUSC.16 It is evident that our present findings provide new evidence supporting this preliminary analysis. In addition to the analysis in lung cancer, TBX5-AS1 is observed to shorten the relapse-free survival of breast cancer patients.29 Based on the evidence above, we concluded that TBX5-AS1 was involved in the progression of NSCLC.
However, the understanding of TBX5-AS1 in cancer still formed at the level of bioinformatics analysis using the existing database16,28,29 and thus further experimental research is still needed. LncRNAs could serve as competitive endogenous RNAs (ceRNAs) by sponging the endogenous suppressive effects of microRNAs or RNA-binding proteins (RBPs) on their targeted transcripts.30,31 Given these biological characteristics of lncRNAs, we validated the effect of TBX5-AS1 on various aspects of cell phenotypes, including survival, proliferation, migration, invasion, and apoptosis. As expected, TBX5-AS1 affected the cell phenotypes in multiple NSCLC cell lines. The overexpression of TBX5-AS1 inhibited the cell viability, colony formation, migration, and invasion, while it promoted the apoptosis. Conversely, knockdown of TBX5-AS1 showed the completely opposite results. To our knowledge, no previous study has investigated the biological functions of TBX5-AS1 on NSCLC at a cellular level. Nevertheless, many other long noncoding antisense RNAs have been explored in NSCLC cells, such as actin filament-associated protein 1 antisense RNA 1 (AFAP1-AS1),32 SBF2 antisense RNA 1 (SBF2-AS1),33 etc. These antisense RNA both were up-regulated in NSCLC and promoted malignant phenotype of NSCLC cells. In contrast, we have shown a unique common suppression pattern of TBX5-AS1 in NSCLC cells, compared with other antisense RNA. For example, ZEB1 as an oncogene drives epithelial-to-mesenchymal transition, thus accordingly lncRNA ZEB1-AS1 also acts as an oncogene by promoting malignant progression.34,35 We speculated that the members of the TBX family and their corresponding antisense RNA have different patterns of expression in NSCLC. TBX5 is reported to have a tumor-suppressing effect in colon cancer and NSCLC cells.14,36 Predictably, overexpression of TBX5-AS1 showed the same tumor-suppressing effect in NSCLC cells. However, the exact mechanism between TBX5-AS1 and TBX5 remains unclear and needed to be elucidated. In short, the present evidently demonstrated that TBX5-AS1 acted as a tumor suppressor in NSCLC, and might have the potential to become an effective therapeutic target.
The elucidation of the downstream regulatory pathway of these lncRNAs contributes to understanding the pathogenic mechanisms of disease progression and improving the therapeutic efficiency for targeted therapy. Except for binding to microRNAs or RBPs, lncRNAs could regulate protein translation and modification directly.37 For instance, the antisense RNA of wilms Tumor 1 Antisense RNA (WT1-AS) could maintain the stability and regulate the translation of WT1 mRNA.37 Thus, we thought that TBX5-AS1 might regulate some certain downstream gene. In previous studies, a number of antisense RNAs has been revealed to affect the cell phenotypes by modulating the PI3K/AKT pathway.38,39 Thus, we investigated the hypothesis that TBX5-AS1 might regulate the PI3K/AKT pathway in NSCLC cells. As results, we found that TBX5-AS1 overexpression suppressed the phosphorylation of PI3K and AKT, indicating that TBX5-AS1 might inactivate the PI3K/AKT pathway. Meanwhile, the addition of PI3K activator reversed the TBX5-AS1-induced inactivation of the PI3K/AKT pathway and affected aggressive phenotypes of NSCLC cells. Although no similar studies have demonstrated a relationship between TBX5-AS1 and the PI3K/AKT signaling pathway, another member of the TBX family named as TBX1 is reported to function as a tumor suppressor by inhibiting the activation of the PI3K/AKT pathway23. Taken together, the results could conclude that TBX5-AS1 inhibited the aggressive phenotypes of NSCLC cells through inactivating the PI3K/AKT pathway.
Conclusion
The present study identified that TBX5-AS1 served as a tumor suppressor in NSCLC. We confirmed the down-regulation of TBX5-AS1 in NSCLC patients and cell lines. Moreover, we creatively confirmed the inhibition effects of TBX5-AS1 on aggressive phenotypes of NSCLC cells, possibly by inhibiting the activation of the PI3K/AKT pathway. The understanding of this novel biological function and mechanism for TBX5-AS1 is conducive to elucidate the pathogenesis of NSCLC and provide a new target for the treatment of NSCLC.
Ethical Statement
All protocols have been approved by the Ethics Committee of Weifang Yidu Center Hospital and performed in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants.
Disclosure
The authors have no conflicts of interest to declare.
References
1. Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi:10.1056/NEJMoa1616288
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
3. Huang CY, Ju DT, Chang CF, Muralidhar Reddy P, Velmurugan BK. A review on the effects of current chemotherapy drugs and natural agents in treating non-small cell lung cancer. Biomedicine (Taipei). 2017;7(4):23. doi:10.1051/bmdcn/2017070423
4. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi:10.1038/nature25183
5. Hirsch FR, Suda K, Wiens J, Bunn PA. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388(10048):1012–1024. doi:10.1016/S0140-6736(16)31473-8
6. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8
7. Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi:10.1146/annurev-genet-120213-092323
8. Shi C, Zhang L, Qin C. Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res Bull. 2017;132:160–169. doi:10.1016/j.brainresbull.2017.03.010
9. Smolle MA, Bauernhofer T, Pummer K, Calin GA, Pichler M. Current insights into long non-coding RNAs (LncRNAs) in prostate cancer. Int J Mol Sci. 2017;18(2):473. doi:10.3390/ijms18020473
10. Wei MM, Zhou GB. Long non-coding RNAs and their roles in non-small-cell lung cancer. Genomics Proteomics Bioinformatics. 2016;14(5):280–288. doi:10.1016/j.gpb.2016.03.007
11. Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–1967. doi:10.1002/ijc.30546
12. Qiu L, Tang Q, Li G, Chen K. Long non-coding RNAs as biomarkers and therapeutic targets: recent insights into hepatocellular carcinoma. Life Sci. 2017;191:273–282. doi:10.1016/j.lfs.2017.10.007
13. Fang C, Wang L, Gong C, Wu W, Yao C, Zhu S. Long non-coding RNAs: how to regulate the metastasis of non-small-cell lung cancer. J Cell Mol Med. 2020;24(6):3282–3291. doi:10.1111/jcmm.15054
14. Yu J, Ma X, Cheung KF, et al. Epigenetic inactivation of T-box transcription factor 5, a novel tumor suppressor gene, is associated with colon cancer. Oncogene. 2010;29(49):6464–6474. doi:10.1038/onc.2010.370
15. Wu CH, Hsu CL, Lu PC, Lin WC, Juan HF, Huang HC. Identification of lncRNA functions in lung cancer based on associated protein-protein interaction modules. Sci Rep. 2016;6:35939. doi:10.1038/srep35939
16. Xiong Y, Zhang X, Lin Z, et al. SFTA1P, LINC00968, GATA6-AS1, TBX5-AS1, and FEZF1-AS1 are crucial long non-coding RNAs associated with the prognosis of lung squamous cell carcinoma. Oncol Lett. 2019;18(4):3985–3993. doi:10.3892/ol.2019.10744
17. Zhang L, Li S, Choi YL, et al. Systematic identification of cancer-related long noncoding RNAs and aberrant alternative splicing of quintuple-negative lung adenocarcinoma through RNA-Seq. Lung Cancer. 2017;109:21–27. doi:10.1016/j.lungcan.2017.04.009
18. Robbins HL, Hague A. The PI3K/Akt pathway in tumors of endocrine tissues. Front Endocrinol (Lausanne). 2015;6:188.
19. Yang SX, Polley E, Lipkowitz S. New insights on PI3K/AKT pathway alterations and clinical outcomes in breast cancer. Cancer Treat Rev. 2016;45:87–96. doi:10.1016/j.ctrv.2016.03.004
20. Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004. doi:10.1038/nrd1902
21. Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016;67:11–28. doi:10.1146/annurev-med-062913-051343
22. Ke M, He Q, Hong D, et al. Leukemia inhibitory factor regulates marmoset induced pluripotent stem cell proliferation via a PI3K/Akt-dependent Tbx3 activation pathway. Int J Mol Med. 2018;42(1):131–140. doi:10.3892/ijmm.2018.3610
23. Wang N, Li Y, Wei J, et al. TBX1 functions as a tumor suppressor in thyroid cancer through inhibiting the activities of the PI3K/AKT and MAPK/ERK pathways. Thyroid. 2019;29(3):378–394. doi:10.1089/thy.2018.0312
24. Borowicz S, Van Scoyk M, Avasarala S, et al. The soft agar colony formation assay. J Vis Exp. 2014;27(92):e51998.
25. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45(8):1895–1910. doi:10.1016/j.biocel.2013.05.030
26. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
27. Yang F, Song Y, Ge L, Zhao G, Liu C, Ma L. Long non-coding RNAs as prognostic biomarkers in papillary renal cell carcinoma. Oncol Lett. 2019;18(4):3691–3697. doi:10.3892/ol.2019.10684
28. Qiao F, Li N, Li W. Integrative bioinformatics analysis reveals potential long non-coding RNA biomarkers and analysis of function in non-smoking females with lung cancer. Med Sci Monit. 2018;24:5771–5778. doi:10.12659/MSM.908884
29. Cun J, Yang Q. Bioinformatics-based interaction analysis of miR-92a-3p and key genes in tamoxifen-resistant breast cancer cells. Biomed Pharmacother. 2018;107:117–128. doi:10.1016/j.biopha.2018.07.158
30. Kim J, Abdelmohsen K, Yang X, et al. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016;44(5):2378–2392. doi:10.1093/nar/gkw017
31. Zhang K, Zhao Z, Yu J, Chen W, Xu Q, Chen L. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J Cell Biochem. 2018;119(7):6045–6056. doi:10.1002/jcb.26802
32. Yu S, Yang D, Ye Y, et al. Long noncoding RNA actin filament-associated protein 1 antisense RNA 1 promotes malignant phenotype through binding with lysine-specific demethylase 1 and repressing HMG box-containing protein 1 in non-small-cell lung cancer. Cancer Sci. 2019;110(7):2211–2225. doi:10.1111/cas.14039
33. Lv J, Qiu M, Xia W, et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:75. doi:10.1186/s13046-016-0352-9
34. Zhang T, Guo L, Creighton CJ, et al. A genetic cell context-dependent role for ZEB1 in lung cancer. Nat Commun. 2016;7:12231. doi:10.1038/ncomms12231
35. Guo Y, Hu Y, Hu M, He J, Li B. Long non-coding RNA ZEB2-AS1 promotes proliferation and inhibits apoptosis in human lung cancer cells. Oncol Lett. 2018;15(4):5220–5226. doi:10.3892/ol.2018.7918
36. Ma R, Yang Y, Tu Q, Hu K. Overexpression of T-box Transcription Factor 5 (TBX5) Inhibits Proliferation and Invasion in Non-Small Cell Lung Carcinoma Cells. Oncol Res. 2017;25(9):1495–1504. doi:10.3727/096504017X14883287513729
37. Zhang Y, Fan L-J, Zhang Y, Jiang J, Qi X-W. Long non-coding wilms tumor 1 antisense RNA in the development and progression of malignant tumors. Front Oncol. 2020;10:35.
38. Zhang JJ, Xu WR, Chen B, et al. The up-regulated lncRNA DLX6-AS1 in colorectal cancer promotes cell proliferation, invasion and migration via modulating PI3K/AKT/mTOR pathway. Eur Rev Med Pharmacol Sci. 2019;23(19):8321–8331. doi:10.26355/eurrev_201910_19143
39. Li Y, Zheng LL, Huang DG, Cao H, Gao YH, Fan ZC. LNCRNA CDKN2B-AS1 regulates mesangial cell proliferation and extracellular matrix accumulation via miR-424-5p/HMGA2 axis. Biomed Pharmacother. 2020;121:109622. doi:10.1016/j.biopha.2019.109622
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.