Back to Journals » OncoTargets and Therapy » Volume 14
LncRNA ST8SIA6-AS1 Promotes Cholangiocarcinoma Progression by Suppressing the miR-145-5p/MAL2 Axis
Authors He J, Yan H, Wei S, Chen G
Received 31 December 2020
Accepted for publication 14 April 2021
Published 17 May 2021 Volume 2021:14 Pages 3209—3223
DOI https://doi.org/10.2147/OTT.S299634
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Junchuang He, Hongxian Yan, Sidong Wei, Guoyong Chen
Department of Hepatobiliary Pancreatic Surgery, Henan Province People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou, 45003, Henan, People’s Republic of China
Correspondence: Junchuang He
Department of Hepatobiliary Pancreatic Surgery, Henan Province People’s Hospital, People’s Hospital of Zhengzhou University, No. 7 Weiwu Road, Jinshui District, Zhengzhou, 450003, Henan, People’s Republic of China
Tel +86 18838097156
Email [email protected]
Background: The tumor-promoting roles of ST8SIA6-AS1 and miR-145-5p have been found in several cancers, but their function in cholangiocarcinoma (CHOL) remains speculative. The purpose of this study was to examine the regulatory functions of the ST8SIA6-AS1/MAL2/miR-145-5p pathway in CHOL progression.
Methods: RT-qPCR assay was used to detect ST8SIA6-AS1 expression in CHOL tissues and cell lines. Cell migration, apoptosis, invasion, and proliferation abilities were assessed by RIP, RNA pull-down, and luciferase assays. CCK-8, BrdU, transwell, and FITC assays to investigate the regulatory functions of ST8SIA6-AS1, miR-145-5p, and MAL2 function in CHOL cells.
Results: Findings revealed the enrichment of ST8SIA6-AS1 in CHOL tissues and cell lines. It was also found that ST8SIA6-AS1 facilitated cell growth and migration, but it reduced the apoptosis level of the CHOL cells. The results of experiments showed that ST8SIA6-AS1 sponged miR-145-5p, thereby allowing MAL2 to exert its biological function on CHOL cells.
Conclusion: This research suggested that the ST8SIA6-AS1/miR-145-5p/MAL2 axis could enhance CHOL progression, which might be useful to improve the clinical outcomes of CHOL patients.
Keywords: ST8SIA6-AS1, miR-145-5p, MAL2, cholangiocarcinoma, proliferation, apoptosis, migration
Introduction
Cholangiocarcinoma (CHOL), the second most commonly diagnosed primary hepatic tumor after hepatocellular carcinoma, originates from the bile ducts. In the last three decades, cases of CHOL have been on the rise, especially in East Asia.1,2 Despite the use of advanced therapeutic methods such as surgery and chemotherapy to treat CHOL patients, the incidence rate and mortality rate of this cancer remain high.3 Therefore, an understanding of the underlying molecular mechanisms of CHOL is needed in improving CHOL prognosis and treatment.
Long non-coding RNAs (lncRNAs) play key roles in the development of malignancy. While lncRNAs cannot code protein, they can regulate gene expression at epigenetic levels.4 Previous research reported that lncRNAs could sponge microRNAs (miRNAs) and regulate the downstream genes.5 Much effort has been made to investigate the regulatory functions of lncRNAs in various cancers, such as osteosarcoma cancer, colorectal cancer, and prostate cancer.6–8 For example, one study found that lncRNA enhanced the invasion abilities of breast cancer cells by activating the Wnt/β-catenin/mTOR/PI3K signaling pathway.9 In another study, lncRNA SNHG5 promoted bladder cancer by targeting p27.10 Moreover, lncRNA FAM83H-AS1 accelerated colorectal carcinoma development by targeting the Notch signaling pathway.11 Research revealed that lncRNAGas5 regulated PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma cells.12 Several studies suggested that lncRNA ST8SIA6-AS1 played an important role in the growth, migration, and invasion of different cancers, such as hepatocellular carcinoma, colorectal cancer, and breast cancer.13–15 However, very little is known about how ST8SIA6-AS1 influences CHOL progression.
MiRNAs are non-coding RNAs with approximately 19–22 nucleotides and the ability to regulate gene expression by suppressing or degrading mRNAs.16 Growing evidence has indicated that miRNAs can regulate the progression of various cancers by regulating cell growth, migration, invasion, and apoptosis, such as pancreatic cancer, papillary thyroid cancer, and ovarian cancer.17–19 For instance, miR-133a-3p repressed the growth and metastasis of gastric cancer cells via blocking autophagy-mediated glutaminolysis.20 MiRNA-106a has been confirmed to promote breast cancer cells’ proliferation, clonogenicity, migration, and invasion but inhibit their apoptosis and chemosensitivity.21 Previous research showed that miRNA‑486‑5p could suppress liver cancer progression by targeting sirt1.37 A decrease in miR-145-5p expression was observed in CHOL cells, which activated the expression of STAT1.22 In addition, miR-145-5p targeted NUAK1 to restrain the carcinogenesis of the CHOL cells.23 It was also reported that the progression of the CHOL cells could be repressed by miR-145-5p via the sirt3/GDH axis.24 However, whether ST8SIA6-AS1 could sponge miR-145-5p and influence CHOL progression remains unclear.
Mal, T cell differentiation protein 2 (MAL2) gene is located on chromosome 8q24.12, and it consists of 4 exons. This gene can encode a multi-span transmembrane protein, which belongs to the MAL proteolipid family.25,26 Various pieces of evidence demonstrated the involvement of MAL2 in accelerating cell growth, migration and invasion in many cancer types, such as breast cancer, papillary thyroid cancer, and hepatocellular carcinoma.27–29 Two studies reported the enrichment of MAL2 expression in CHOL tissues and cell lines.30,31 However, the tumorigenic role of the ST8SIA6-AS1/miR-145-5p/MAL2 axis in CHOL needs to be further explored.
This study aimed to examine the regulatory impacts of ST8SIA6-AS1 on the miR-145-5p/MAL2 axis in CHOL cells. We hypothesized that ST8SIA6-AS1 could promote CHOL progression by suppressing the miR-145/5p-MAL2 axis.
Materials and Methods
Bioinformatics Analysis
The ENCORI (The Encyclopedia of RNA Interactomes, http://starbase.sysu.edu.cn/) algorithm was used to analyze the correlation between ST8SLA6-AS1 and CHOL progression. GSE77984 was the mRNA expression profile, and it included CHOL tissues and normal human cholangiocyte tissues. The upregulated differentially expressed genes (DEGs) were screened from GSE77984 with the selection criteria of adjusted P<0.05 and logFC>3. The target genes of miR-145-5p were predicted by ENCORI. The common genes were overlapped from GSE77984, and the predicted target genes were analyzed using Venny 2.1.0. Finally, the TCGA database was used to show the common genes in CHOL.
Patient Samples, Cell Lines, and Cell Transfection
A total of 35 CHOL specimens and paracancerous tissues were obtained from 35 patients at Henan Province People’s Hospital, People’s Hospital of Zhengzhou University, China (No. KY2018-099-03, 2018-5-29). Informed consent forms were completed by the patients, and the survey was approved by the Ethics Committee of this hospital. The patients’ clinical characteristics are presented in Table 1.
 |
Table 1 The Clinical Characteristics of 35 Cholangiocarcinoma Patients |
Human CHOL cell lines (TFK-1, CCLP, RBE, and HUCCT1) and the normal HIBEC epithelial cell were purchased from ATCC (Manassas, VA, USA). TFK-1, CCLP, and HUCCT1 cells were seeded in DMEM (Gibco, USA), while RBE and HIBEC cells were seeded in the RPMI-1640 medium (Gibco, USA). All the culture media contained 10% FBS (Gibco, USA). The cells collected from patients were cultured at 37 °C in an atmosphere containing 5% CO2. siRNA-ST8SIA6-AS1 (si-lnc), miR-145-5p mimics, inhibitor, siRNA-MAL2, and negative control (NC) were obtained from RiboBio (Guangzhou, China). The cells were transfected for 48 h using Lipofectamine 2000 Transfection Reagent (Invitrogen, MA, USA). The non-transfected cells were regarded as the blank group.
Nucleic Acid Isolation and RT-qPCR
The MAL2 mRNA and ST8SIA6-AS1 lncRNA were isolated using TRIzol Reagent (Cat#: 15596018, Thermo, USA). The miRcute miRNA Isolation Kit (DP501, Tiangen, China) was employed to isolate and purify the miRNAs. The separation of the nuclear and cytosolic fractions was conducted using the Ambion PARIS Kit (AM1921, Life, USA). Then, the cDNA of MAL2 and ST8SIA6-AS1 was obtained using the PrimeScript First Strand cDNA Synthesis Kit (RR037A, Takara, China), while that of miR-145-5p was obtained using the miRcute miRNA First-strand cDNA Synthesis Kit (KR211, Tiangen, China). Finally, the expression of MAL2 and ST8SIA6-AS1 was identified using SYBR Premix Ex Taq (RR420A, Takara, China), while the expression of miR-145-5p was observed using the miRcute-enhanced miRNA Fluorescence Quantitative Detection Kit (FP411, Tiangen, China). GAPDH was used as the internal control for MAL2, while U6 was employed as the internal control for miR-145-5p. The gene expression was determined using the of 2−ΔΔCt technique. The primer sequences are listed in Table 2.
 |
Table 2 Primer Sequences |
CCK-8 Assay
CCLP and RBE cells (5×103) were cultured in 96-well plates. The cell viability was detected after 24 h, 48 h, 72 h, and 96 h. 10 µL CCK-8 solution (Cat#: K1018; APExBIO, China) was added to each well, and the mixture was incubated for 2 h. The optical density (OD) value at 450 nm was obtained with a multimode-plate-reader (Thermo, USA).
BrdU Assay
CCLP and RBE cells (5×103) were cultured in 96-well plates. The cells were incubated with BrdU (Cat#: ab126556, Abcam, USA) for 4 h. Then, cells were permeabilized and treated with the BrdU antibody and incubated for 2 h. Finally, the cells were treated with the anti-mouse antibody for 1 h. The OD value (450 nm) was obtained with a multimode-plate-reader (Thermo, USA).
Transwell Assay
The 8 μm pore size chamber (Cat#: #3244, Corning, NY, USA) in the 24-well plate was used to carry out cell invasion measurement. The lower chamber was pre-coated with matrigel (Sigma, USA) and placed in 10% FBS medium, while the upper chamber was cultured with 8×104 CCLP and RBE cell suspensions in a serum-free medium. After incubating the cells for 48 h, the cells at the top layer were removed, and the cells at the bottom layer were fixed with methanol and stained with 0.1% crystal violet. Finally, five random fields from each chamber were photographed under the microscope (Olympus, Tokyo, Japan).
Apoptosis Assay
The apoptosis level of the CCLP and RBE cells was measured using the Annexin V-FITC/PI Apoptosis Detection Kit (Cat#: 556547; BD, USA). After the cells were collected and washed, they were suspended in the binding buffer with 5 µL FITC plus 5 µL PI. After that, the cells were incubated for 20 min in the absence of light. Next, they were washed twice and suspended in 400 µL binding buffer. The cell apoptosis rate was finally determined using the FACSAria flow cytometer (BD, USA).
Dual-Luciferase Reporter Assay
The psiCHECK2 ST8SIA6-AS1 wild-type or mutated vector, and psiCHECK2 MAL2 wild-type or mutated vector were purchased from Touran Bio (Shanghai, China), and the CCLP and RBE cells were co-transfected with the miR-145-5p mimic or NC using Lipofectamine 3000 Transfection Reagent (Invitrogen, MA, USA) for 48 h. Afterward, the relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Cat#: E1910, Promega, USA).
RIP Assay
This assay was conducted using the EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Cat#: #17-701, sigma, USA). The cell lysates were incubated with magnetic beads conjugated with miR-145-5p mimic or NC, and IgG (Cat#: ab172730, Abcam, UK) or Ago2 (Cat#: ab186733, Abcam, UK) overnight at 4 °C. After immunoprecipitated RNA was purified, RT-qPCR was applied to detect ST8SIA6-AS1 expression.
RNA-Pull Down Analysis
The CCLP and RBE cells were treated with biotin-labeled miR-145-5p (bio- miR-145-5p) and miR-negative control (bio-NC) (RiboBio, Guangzhou, China). After 48 h, the cell lysates were incubated overnight at 4°C with the streptavidin beads (Cat#: #88817, Thermo, USA). Then, the bounded RNAs were eluted and purified with the RNeasy Mini Kit (Cat#: 74104, QIAGEN, Germany). RT-qPCR was finally applied to detect the enrichment of MAL2.
Statistical Analysis
GraphPad Prism 8.0 (GraphPad Prism, USA) was employed for data analysis. Data were presented in the form of mean ± standard deviation (SD). The statistical significance between two groups was obtained using the Wilcoxon test, while the statistical significance among multiple groups was estimated using the one-way ANOVA with Dunnett’s post hoc. The associations between ST8SIA6-AS1 and miR-145-5p expressions, or MAL2 and miR-145-5p expressions were estimated using Pearson correlation analysis. P< 0.05 was considered statistically significant.
Results
The miR-145-5p-MAL2 Axis Was Predicted as the Downstream of ST8SIA6-AS1 in CHOL
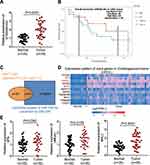
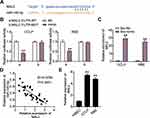
ST8SIA6-AS1 has been reported to promote the development of breast cancer,15,32 hepatocellular cancer,13,33 and colorectal cancer.14 However, its impacts on CHOL cell lines remain unknown. This study examined the expression pattern of ST8SIA6-AS1 in CHOL tissue samples and found that it was significantly upregulated more in CHOL tissues than in adjacent tissues (Figure 1A). After analyzing its expression and survival outcomes of patients with CHOL, it was found that from month 13 to month 53, patients with a higher ST8SIA6-AS1 expression level had a low survival rate compared to those with a lower ST8SIA6-AS1 expression level (Figure 1B). Based on this finding, it was hypothesized that ST8SIA6-AS1 could enhance CHOL progression. Using the ENCORI algorithm, we identified five potential downstream miRNA targets of ST8SIA6-AS1: miR-142-3p, miR-651-5p, miR-338-3p, miR-145-5p and miR-5195-3p. Among the five miRNAs, we noticed that miR-145-5p functioned as a cancer inhibitor in CHOL cells,22 whereas the effects of the other four miRNAs on CHOL have not been reported. The intersect of the predicted targets of miR-145-5p was found after performing ENCORI and DEG analyses with the selection criteria of adjusted P<0.05 and logFC>3. A total of 11 genes were identified (Figure 1C). The TCGA database showed that the expression levels of SFN, HMGA1 and MAL2 in CHOL were upregulated (Figure 1D). After applying qRT-PCR, MAL2 was found to be highly expressed in CHOL cells (Figure 1E). For this reason, we chose MAL2 as the potential downstream effector of miR-145-5p that could be targeted by ST8SIA6-AS1.
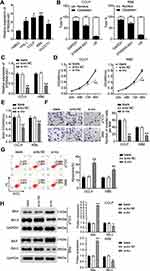
ST8SIA6-AS1 Silence Inhibits the Progression of the CHOL Cells
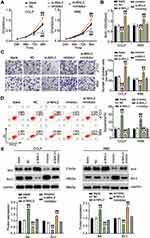
We examined how ST8SIA6-AS1 functioned in the progression of the CHOL cells and found that ST8SIA6-AS1 expression was remarkably upregulated in CHOL cell lines (TFK-1, CCLP, RBE, and HUCCT1) compared with the normal HIBEC epithelial cell. We used CCLP and RBE in subsequent experiments due to the higher expression of ST8SIA6-AS1 in CCLP and RBE cells (Figure 2A). The nucleus–cytoplasm fraction analysis demonstrated that ST8SIA6-AS1 was mainly expressed in the cell cytoplasm (Figure 2B). To explore the function of ST8SIA6-AS1 in CHOL cells, we transfected siRNA-ST8SIA6-AS1 (si-lnc) into CCLP and RBE cells. The expression of ST8SIA6-AS1 was reduced by about 60% in the si-lnc groups compared to the blank control group (Figure 2C). It was also found that the viability levels of the si-lnc groups were lower than those of the blank control group in both CCLP and RBE cells (Figure 2D). Similarly, the proliferation levels in the si-lnc groups declined by 30% reduced compared to the blank control group in both CCLP and RBE cells (Figure 2E). Besides, a decrease in the invasion level of the si-lnc group was discovered compared with the blank control group (Figure 2F). Additionally, the si-lnc groups demonstrated a 2-fold increase in cell apoptosis compared to the blank control group (Figure 2G). Consistently, the Western blot results demonstrated that ST8SIA6-AS1 silence increased BAX expression and reduced Bcl-2 expression in both CCLP and RBE cells (Figure 2H). In sum, our results confirmed that ST8SIA6-AS1 could facilitate CHOL cells’ proliferation and migration, but it could suppress the apoptosis of the CHOL cells.
ST8SIA6-AS1 Interacts with miR-145-5p in CHOL Cells
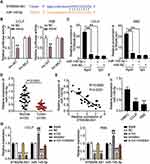
Previous research has demonstrated that lncRNAs can orchestrate miRNA expression and thereby influence tumor progression. The binding site sequence between ST8SIA6-AS1 and miR-145-5p was predicted by starBase (Figure 3A). The luciferase activity results showed that the co-transfection of miR-145-5p mimics and psiCHECK2 ST8SIA6-AS1-wide type (WT) downregulated luciferase activities by about 50% compared to cells transfected with psiCHECK2 ST8SIA6-AS1 mutant (Mut) in both CCLP and RBE cells, meaning miR-145-5p could directly interact with ST8SIA6-AS1 (Figure 3B). Next, the RIP experiment results indicated that ST8SIA6-AS1 interacted with miR-145-5p (Figure 3C). We then assessed the expression level of miR-145-5p in CHOL tissues and found that ST8SIA6-AS1 was downregulated by 70% in CHOL tissues compared to the normal tissues (Figure 3D). A negative correlation between the miR-145-5p expression and ST8SIA6-AS1 expression level in CHOL tissues was also observed (Figure 3E). Furthermore, the expression of miR-145-5p in CCLP and RBE cells declined by 50% compared to the normal HIBEC cell (Figure 3F).
Given the potential interaction between miR-145-5p and ST8SIA6-AS1, we transfected siRNA-ST8SIA6-AS1 (si-lnc) and the miR-145-5p inhibitor in CCLP and RBE cells to confirm the relationship between ST8SIA6-AS1 and miR-145-5p. As showed in Figure 3G, miR-145-5p level in the si-lnc group increased by 1.5-fold, while ST8SIA6-AS1 level in the si-lnc group declined by 70% compared with the blank control group. The miR-145-5p inhibitor group downregulated miR-145-5p expression levels by 70% compared with the blank control group; however, the miR-145-5p inhibitor did not affect the ST8SIA6-AS1 level. In addition, the si-lnc+inhibitor group downregulated ST8SIA6-AS1 level by 70% compared to the blank control group, but no difference was observed in the miR-145-5p levels. Collectively, the results indicated that ST8SIA6-AS1 could sponge miR-145-5p in CHOL cells.
ST8SIA6-AS1 Sponges miR-145-5p Expression to Promote CHOL Progression
We examined whether ST8SIA6-AS1 could influence the CHOL cells by regulating miR-145-5p in CHOL cells. Experimental findings revealed that the miR-145-5p inhibitor accelerated the viability ability of the CHOL cells compared to the blank control group; however, si-lnc counteracted the effect in CCLP and RBE cells (Figure 4A). The miR-145-5p inhibitor group enhanced the proliferation abilities of the CHOL cells by 30% compared to the blank control group; however, si-lnc reversed the effect in the tumor cells (Figure 4B). Moreover, the miR-145-5p inhibitor group also enhanced the invasion ability of the CHOL cells compared with the blank control group; nonetheless, this enhancement was reversed by si-lnc (Figure 4C). Finally, the apoptosis rate of the miR-145-5p inhibitor group was repressed by 50% with the blank control group; however, while this repressive effect was counteracted by si-lnc (Figure 4D). The expression levels of BAX and Bcl-2 further confirmed that ST8SIA6-AS1 silence could attenuate the repressive effect of the miR-145-5p inhibitor on CCLP and RBE cells’ apoptosis (Figure 4E). Overall, these results demonstrated that by sponging miR-145-5p, ST8SIA6-AS1 could enhance CHOL progression.
MiR-145-5p Targets MAL2 and Inhibits the Expression of MAL2 in CHOL Cells
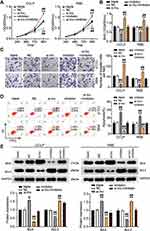
We further confirmed the impacts of miR-145-5p on CHOL development. As demonstrated in Figure 5A, the binding site sequence between MAL2 and miR-145-5p was analyzed by starBase. The miR-145-5p mimics transfected with the psiCHECK2 MAL2 3ʹUTR WT plasmid decreased luciferase activities by 50%, but no changes were found in the psiCHECK2 MAL2 3ʹUTR Mut plasmid, meaning miR-145-5p could directly interact with MAL2 (Figure 5B). The RNA pull-down assay results indicated that miR-145-5p interacted with MAL2 in the CCLP and RBE cells (Figure 5C). In addition, a negative correlation between the expression of miR-145-5p and MAL2 was found in CHOL tissues (Figure 5D). Meanwhile, MAL2 expression in CCLP and RBE cells was upregulated by 5-fold compared to normal HIBEC cells (Figure 5E). In short, our results suggested that miR-145-5p could directly target and repress MAL2 in CHOL cells.
MiR-145-5p Regulates the Progression of the CHOL Cells by Inhibiting MAL2
We treated CCLP and RBE cells with si-MAL2 and the miR-145-5p inhibitor to examine the impacts of miR-145-5p on MAL2 in CHOL cells. Our findings revealed that si-MAL2 group had a lower cell viability level compared to the blank control group, while the miR-145-5p inhibitor group demonstrated had a higher cell viability level, which was abrogated by si-MAL2 co-transfection in the CCLP and RBE cells (Figure 6A). Additionally, the si-MAL2 group inhibited cell proliferation by 30% compared with the blank control group, while the cell proliferation level in the miR-145-5p inhibitor group increased by more than 30%. This effect was reversed in si-MAL2 +inhibitor groups (Figure 6B). Besides, the si-MAL2 group suppressed the invasion ability of CCLP cells by 75% and that of RBE cells by 50% compared with the blank control group, while an increase in cell invasion was observed in the inhibitor group. The si-MAL2+inhibitor groups reversed the effect in both CCLP and RBE cells (Figure 6C). Furthermore, the apoptosis rate of the si-MAL2 group was enhanced by 2.5-fold compared with the blank control group, while the apoptosis rate of the miR-145-5p inhibitor group was reduced compared with the blank control group. This effect was abrogated in the CCLP and RBE cells transfected with the si-MAL2 +inhibitor (Figure 6D). Figure 6E indicated the expression of two typical apoptosis-related factors: BAX and Bcl-2 in CCLP and RBE cells after transfection. As shown in the Figure 6E, MAL2 silence could increase the BAX expression level and reduce the Bcl-2 expression level, while the miR-145-5p inhibitor produced an opposite result. When the CHOL cells were co-treated with the si-MAL2 +inhibitor, this effect on the expression of BAX and Bcl-2 was abrogated. Overall, our findings indicated that miR-145-5p could repress CHOL cells’ proliferation and migration but improve CHOL cells’ apoptosis by targeting MAL2.
Discussion
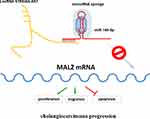
This study revealed that ST8SIA6-AS1 expression was upregulated in CHOL tissues and cell lines. We found that ST8SIA6-AS1 improved the proliferation and migration abilities of the CHOL cells, but it inhibited the apoptosis rate of the CHOL cells by sponging miR-145-5p. It was discovered that ST8SIA6-AS1 inhibited the tumor suppressor impacts of miR-145-5p on MAL2. A proposed model showing the molecular mechanism of ST8SIA6-AS1 in CHOL is presented in Figure 7. Therefore, the ST8SIA6-AS1/miR-145-5p/MAL2 axis could be considered a novel treatment pathway for CHOL.
Over the years, much effort has been made in exploring the ability of lncRNAs to regulate human malignancies. Research has reported the involvement of lncRNA ST8SIA6-AS1 in the development of several tumors. For example, silenced ST8SIA6-AS1 restrained the viability, migration, and invasion abilities of hepatocellular carcinoma cells, but it boosted the apoptosis rate of hepatocellular carcinoma cells via the miR-5195-3p/HOXB6 axis.13 Besides, ST8SIA6-AS1 knockdown reduced cell growth, migration, and invasion via the miR-5195/PCBP2 axis in colorectal cancer cells.14 Another study indicated that ST8SIA6-AS1 overexpression elevated the proliferation, invasion, and migration abilities of breast cancer cells by activating the p38 MAPK signaling pathway.15 In this study, we researched the role of ST8SIA6-AS1 in CHOL cells and found that ST8SIA6-AS1 expression was enhanced in CHOL tissues and cell lines. More specifically, we noticed that the downregulation of ST8SIA6-AS1 repressed the growth and migration level of CCLP and RBE cells, while it promoted the apoptosis rate of CCLP and RBE cells. Moreover, by sponging miR-145-5p, ST8SIA6-AS1 counteracted the tumor suppressor effect of miR-145-5p on CHOL cells.
Several studies reported that miRNAs participated in cancer occurrence and progression by interacting lncRNAs.35–37 Zhou et al revealed that miR-145-5p suppressed the proliferation, migration and invasion of gastric cancer epithelial cells via the ANGPT2/NOD_LIKE_RECEPTOR axis.39 Additionally, the loss of miR-145-5p caused ceruloplasmin interference with the PHD-Iron axis and HIF-2α stabilization in lung adenocarcinoma-mediated angiogenesis.40 In CHOL cells, Zeng et al clarified that lncRNA TUG1 sponged miR-145-5p to promote intrahepatic CAA progression and regulate glutamine metabolism via the sirt3/GDH axis.24 Xiong et al indicated that miR-145-5p suppressed cancer-associated adipocytes (CAA)’s proliferation and invasion abilities by targeting NUAK1 and activating Akt signaling.23 Similarly, we noticed a decrease of miR-145-5p expression in CHOL tissues and cell lines, which was negatively correlated with the ST8SIA6-AS1 level or MAL2 level in CHOL tissues. Furthermore, it was found that miR-145-5p repressed cell growth and promoted cell apoptosis in CHOL by inhibiting MAL2; however, ST8SIA6-AS1 inhibited miR-145-5p expression, which further facilitated the progression of CHOL.
Recent findings have improved the understanding of the role of MAL2 in a variety of cancers, including oral squamous cell carcinoma and breast cancer.27,41 Evidence in the literature showed that MAL2 was overexpressed in breast cancer cell lines. The downregulation of MAL2 inhibited the proliferation, migration, and invasion abilities of breast cancer cell lines.27 Additionally, lncRNA RP11‐284F21.9 facilitated oral squamous cell carcinoma development via the miR‐383‐5p/MAL2 axis. The knockdown of RP11‐284F21.9 could reduce the expression of MAL2 and suppress the proliferation, migration, and invasion levels of oral squamous cell carcinoma cell lines.41 Two studies suggested that MAL2 expression was elevated in CHOL tissues and cell lines; however, the specific mechanism was not clear.30,31 Our study also demonstrated that MAL2 expression was elevated in CHOL tissues and cell lines. We conducted several experiments that indicated the knockdown of MAL2 in CCLP and RBE cells. The downregulation of MAL2 significantly restrained the proliferation and migration abilities of the CHOL cells, but it increased the apoptosis of the CHOL cells. Moreover, miR-145-5p inhibited MAL2 expression, which further suppressed CHOL progression.
Despite the usefulness of our findings, this research is not immune to a few limitations. Previous evidence showed that MAL2 was inhibited by miR-216b-5p, which further reduced chronic constriction injury-induced neuropathic pain in female rats via the Wnt/β-catenin signaling pathway. In the future, the signaling pathways involved in the ST8SIA6-AS1/miR-145-5p/MAL2 axis in CHOL progression should be explored. Besides, this research only investigated the effect of the ST8SIA6-AS1/miR-145-5p/MAL2 axis on CHOL in vitro. In the future, in vivo experiments should be performed to verify the conclusions of this study.
Conclusion
In sum, this research suggested that ST8SIA6-AS1 could facilitate CHOL cells’ proliferation and migration, but it could repress CHOL cells’ apoptosis through the miR-145-5p/MAL2 axis. The knowledge of this research might be instrumental in improving the clinical outcomes of CHOL patients.
Abbreviations
CHOL, cholangiocarcinoma; lncRNAs, long non-coding RNAs; miRNAs, microRNAs; MAL2, Mal, T cell differentiation protein 2; DEGs, differentially expressed genes; NC, negative control; WT, wide type; Mut, mutant; RIP, RNA Immunoprecipitation.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
The present study was approved by the Ethics Committee of Henan Province People’s Hospital, People’s Hospital of Zhengzhou University. The processing of clinical tissue samples is in strict compliance with the ethical standards of the Declaration of Helsinki. All patients signed written informed consent.
Consent for Publication
Consent for publication was obtained from the participants.
Funding
This manuscript was supported by Joint Co-construction Project of Henan Medical Science and Technology Research Plan in 2020 (LHGJ20200010).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15(2):95–111. doi:10.1038/nrclinonc.2017.157
3. Schroeder H, Boyet S, Nehlig A. Effects of caffeine and doxapram perfusion on local cerebral glucose utilization in conscious rats. Eur J Pharmacol. 1989;167(2):245–254. doi:10.1016/0014-2999(89)90585-2
4. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi:10.1038/nrg.2015.10
5. Dutta A, Roy A, Chatterjee S. Long noncoding RNAs in cancer immunity: a new avenue in drug discovery. Drug Discov Today. 2020;26:264–272. doi:10.1016/j.drudis.2020.07.026
6. Zhou Y, Feng D, Gu X, Gao A, Liu Y. The role and clinical significance of long noncoding RNA zinc finger E-box-binding homeobox two antisense RNA 1 in promoting osteosarcoma cancer cell proliferation, inhibiting apoptosis and increasing migration by regulating miR-145. Anticancer Drugs. 2020.
7. Li W, He Y, Cheng Z. Long noncoding RNA XIST knockdown suppresses the growth of colorectal cancer cells via regulating microRNA-338-3p/PAX5 axis. Eur J Cancer Prev. 2020.
8. Chen QH, Li B, Liu DG, Zhang B, Yang X, Tu YL. LncRNA KCNQ1OT1 sponges miR-15a to promote immune evasion and malignant progression of prostate cancer via up-regulating PD-L1. Cancer Cell Int. 2020;20:394. doi:10.1186/s12935-020-01481-8
9. Liu L, Yu D, Shi H, Li J, Meng L. Reduced lncRNA Aim enhances the malignant invasion of triple-negative breast cancer cells mainly by activating Wnt/β-catenin/mTOR/PI3K signaling. Die Pharmazie. 2017;72(10):599–603. doi:10.1691/ph.2017.7547
10. Ma Z, Xue S, Zeng B, Qiu D. lncRNA SNHG5 is associated with poor prognosis of bladder cancer and promotes bladder cancer cell proliferation through targeting p27. Oncol Lett. 2018;15(2):1924–1930. doi:10.3892/ol.2017.7527
11. Lu S, Dong W, Zhao P, Liu Z. lncRNA FAM83H-AS1 is associated with the prognosis of colorectal carcinoma and promotes cell proliferation by targeting the Notch signaling pathway. Oncol Lett. 2018;15(2):1861–1868. doi:10.3892/ol.2017.7520
12. Zhang XF, Ye Y, Zhao SJ. LncRNA Gas5 acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma. Oncotarget. 2018;9(3):3519–3530. doi:10.18632/oncotarget.23336
13. Li Y, Jiang A. ST8SIA6-AS1 promotes hepatocellular carcinoma by absorbing miR-5195-3p to regulate HOXB6. Cancer Biol Ther. 2020;21(7):647–655. doi:10.1080/15384047.2020.1743150
14. Huang CM, Cao GY, Yang CX, et al. LncRNA ST8SIA6-AS1 promotes colorectal cancer cell proliferation, migration and invasion by regulating the miR-5195/PCBP2 axis. Eur Rev Med Pharmacol Sci. 2020;24(8):4203–4211. doi:10.26355/eurrev_202004_21000
15. Fang K, Hu C, Zhang X, et al. LncRNA ST8SIA6-AS1 promotes proliferation, migration and invasion in breast cancer through the p38 MAPK signalling pathway. Carcinogenesis. 2020;41(9):1273–1281. doi:10.1093/carcin/bgz197
16. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi:10.1146/annurev-pathol-012513-104715
17. Xu X, Zheng S. MiR-887-3p negatively regulates STARD13 and promotes pancreatic cancer progression. Cancer Manag Res. 2020;12:6137–6147. doi:10.2147/CMAR.S260542
18. Zhang W, Ji W, Li T, Liu T, Zhao X. MiR-145 functions as a tumor suppressor in papillary thyroid cancer by inhibiting RAB5C. Int J Med Sci. 2020;17(13):1992–2001. doi:10.7150/ijms.44723
19. He F, Zu D, Lan C, Niu J, Nie X. hsa-microRNA-411-5p regulates proliferation, migration and invasion by targeting the hyaluronan mediated motility receptor in ovarian cancer. Exp Ther Med. 2020;20(3):1899–1906. doi:10.3892/etm.2020.8899
20. Zhang X, Li Z, Xuan Z, et al. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J Exp Clin Cancer Res. 2018;37(1):320. doi:10.1186/s13046-018-0993-y
21. You F, Luan H, Sun D, et al. miRNA-106a promotes breast cancer cell proliferation, clonogenicity, migration, and invasion through inhibiting apoptosis and chemosensitivity. DNA Cell Biol. 2019;38(2):198–207. doi:10.1089/dna.2018.4282
22. Goeppert B, Truckenmueller F, Ori A, et al. Profiling of gallbladder carcinoma reveals distinct miRNA profiles and activation of STAT1 by the tumor suppressive miRNA-145-5p. Sci Rep. 2019;9(1):4796. doi:10.1038/s41598-019-40857-3
23. Xiong X, Sun D, Chai H, et al. MiR-145 functions as a tumor suppressor targeting NUAK1 in human intrahepatic cholangiocarcinoma. Biochem Biophys Res Commun. 2015;465(2):262–269. doi:10.1016/j.bbrc.2015.08.013
24. Zeng B, Ye H, Chen J, et al. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget. 2017;8(69):113650–113661. doi:10.18632/oncotarget.21922
25. Marazuela M, Alonso MA. Expression of MAL and MAL2, two elements of the protein machinery for raft-mediated transport, in normal and neoplastic human tissue. Histol Histopathol. 2004;19(3):925–933. doi:10.14670/HH-19.925
26. Marazuela M, Acevedo A, García-López MA, Adrados M, de Marco MC, Alonso MA. Expression of MAL2, an integral protein component of the machinery for basolateral-to-apical transcytosis, in human epithelia. J Histochem Cytochem. 2004;52(2):243–252. doi:10.1177/002215540405200212
27. Bhandari A, Shen Y, Sindan N, et al. MAL2 promotes proliferation, migration, and invasion through regulating epithelial-mesenchymal transition in breast cancer cell lines. Biochem Biophys Res Commun. 2018;504(2):434–439. doi:10.1016/j.bbrc.2018.08.187
28. Gao X, Chen Z, Li A, Zhang X, Cai X. MiR-129 regulates growth and invasion by targeting MAL2 in papillary thyroid carcinoma. Biomed Pharmacother. 2018;105:1072–1078. doi:10.1016/j.biopha.2018.06.050
29. de Marco MC, Puertollano R, Martínez-Menárguez JA, Alonso MA. Dynamics of MAL2 during glycosylphosphatidylinositol-anchored protein transcytotic transport to the apical surface of hepatoma HepG2 cells. Traffic. 2006;7(1):61–73. doi:10.1111/j.1600-0854.2005.00361.x
30. López-Coral A, Del Vecchio GJ, Chahine JJ, Kallakury BV, Tuma PL. MAL2-induced actin-based protrusion formation is anti-oncogenic in hepatocellular carcinoma. Cancers (Basel). 2020;12(2):422. doi:10.3390/cancers12020422
31. Yoo HJ, Yun BR, Kwon JH, et al. Genetic and expression alterations in association with the sarcomatous change of cholangiocarcinoma cells. Exp Mol Med. 2009;41(2):102–115. doi:10.3858/emm.2009.41.2.013
32. Jeong G, Bae H, Jeong D, et al. A Kelch domain-containing KLHDC7B and a long non-coding RNA ST8SIA6-AS1 act oppositely on breast cancer cell proliferation via the interferon signaling pathway. Sci Rep. 2018;8(1):12922. doi:10.1038/s41598-018-31306-8
33. Fei Q, Song F, Jiang X, et al. LncRNA ST8SIA6-AS1 promotes hepatocellular carcinoma cell proliferation and resistance to apoptosis by targeting miR-4656/HDAC11 axis. Cancer Cell Int. 2020;20:232. doi:10.1186/s12935-020-01325-5
34. Dai F, Liu T, Zheng S, et al. MiR-106b promotes migration and invasion through enhancing EMT via downregulation of Smad 7 in Kazakh’s esophageal squamous cell carcinoma. Tumour Biol. 2016;37(11):14595–14604. doi:10.1007/s13277-016-5338-x
35. Liu C, Kelnar K, Liu B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–215. doi:10.1038/nm.2284
36. Hwang WL, Jiang JK, Yang SH, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16(3):268–280. doi:10.1038/ncb2910
37. Yan X, Liu X, Wang Z, et al. MicroRNA‑486‑5p functions as a tumor suppressor of proliferation and cancer stem‑like cell properties by targeting Sirt1 in liver cancer. Oncol Rep. 2019;41(3):1938–1948. doi:10.3892/or.2018.6930
38. Zhou K, Song B, Wei M, Fang J, Xu Y. MiR-145-5p suppresses the proliferation, migration and invasion of gastric cancer epithelial cells via the ANGPT2/NOD_LIKE_RECEPTOR axis. Cancer Cell Int. 2020;20:416. doi:10.1186/s12935-020-01483-6
39. Tsai YM, Wu KL, Chang YY, et al. Loss of miR-145-5p causes ceruloplasmin interference with PHD-iron axis and HIF-2α stabilization in lung adenocarcinoma-mediated angiogenesis. Int J Mol Sci. 2020;21(14):5081. doi:10.3390/ijms21145081
40. Shao B, Fu X, Li X, Li Y, Gan N. RP11-284F21.9 promotes oral squamous cell carcinoma development via the miR-383-5p/MAL2 axis. J Oral Pathol Med. 2020;49(1):21–29. doi:10.1111/jop.12946
41. Fan X, Bian W, Liu M, Li J, Wang Y. MiR-216b-5p attenuates chronic constriction injury-induced neuropathic pain in female rats by targeting MAL2 and inactivating Wnt/β-catenin signaling pathway. Neurochem Int. 2020;104930. doi:10.1016/j.neuint.2020.104930
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.