Back to Journals » International Journal of Women's Health » Volume 11
LncRNA SRA1 may play a role in the uterine leiomyoma tumor growth regarding the MED12 mutation pattern
Authors Akbari M , Yassaee F, Aminbeidokhti M, Abedin-Do A , Mirfakhraie R
Received 12 April 2019
Accepted for publication 1 August 2019
Published 29 August 2019 Volume 2019:11 Pages 495—500
DOI https://doi.org/10.2147/IJWH.S211632
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Elie Al-Chaer
Mojdeh Akbari,1 Fakhrolmolouk Yassaee,2 Mona Aminbeidokhti,1 Atieh Abedin-Do,1,3 Reza Mirfakhraie1,4
1Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 2Department of Obstetrics and Gynecology, Taleghani Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 3GREB, Dental Faculty, Department of Regenerative Medicine, Faculty of Medicine, Laval University, Quebec, Canada; 4Department of Molecular Genetics, Genomic Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Correspondence: Reza Mirfakhraie
Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Koodakyar St, Velenjak Ave, Chamran highway, Tehran 19395-4719, Iran
Tel/Fax +98 212 387 2572
Email [email protected]
Fakhrolmolouk Yassaee
Department of Obstetrics and Gynecology, Taleghani Hospital, Shahid Beheshti University of Medical Sciences, PO Box 1985717413, Tehran, Iran
Email [email protected]
Background: Uterine leiomyomas (ULMs) are benign uterine tumors that are estrogen-dependent. Recent studies suggest that the abnormal expression of the steroid receptor RNA activator 1 (SRA1) long non-coding RNA (lncRNA) might participate in the mechanisms of tumorigenesis of some hormone-dependent tumors including breast cancer. SRA1 is known to enhance the transcriptional activity of steroid receptors and also promotes steroidogenesis. The level of steroid hormones, such as estrogen and the progesterone, and their receptors play an important role in the development and growth of leiomyoma. The aim of the present study was to determine the expression level of lncRNA SRA1 in ULM tissues considering the MED12 mutation pattern.
Methods: Mutation screening was performed for MED12 exons 1 and 2 and the intronic flanking regions using Sanger sequencing in 60 ULM tissues. Quantitative real-time polymerase chain reaction (qRT-PCRs) was performed in order to estimate the expression of lncRNA SRA1 in leiomyoma samples with and without MED12 gene mutations. The expression results were analyzed by using LinReg and REST software.
Results: Mutations were detected in exon 2 of the MED12 in 28 (46.67%) ULM samples; including, 21 (75%) missense mutations and 7 (25%) in-frame deletions. No mutation was detected in the MED12 exon 1. LncRNA SRA1 was over-expressed in ULM samples without MED12 mutation compared with ULM samples harboring MED12 mutation (Expression ratio=2.5, P-value=0.004).
Conclusion: Present results suggest that lncRNA SRA1 may explain the phenotypic difference observed in the tumor size of ULM samples considering MED12 mutation pattern. Therefore, it serves as a good therapeutic target and provides new insight into understanding the disease molecular mechanism.
Keywords: MED12, mutation, SRA1, lncRNA, uterine leiomyoma
Introduction
Uterine leiomyomas (ULMs) (also named as fibroids) are the most common type of women’s health disorders. According to statistical information, 40–80% of reproductive-aged women are diagnosed with ULMs.1 These benign tumors arise due to monoclonal transformation of the myometrium, smooth muscle cell layer of the uterus. They can cause several complications such as infertility, miscarriages, bleeding, pain, dysmenorrhea, menorrhagia, and dyspareunia. Excluding hysterectomy, most treatments often provide only temporary relief and are not successful in all patients; therefore, its social impact is significant, with money expend on medical support as well as the obvious impact on women’s productivity and work absenteeism.2 Both environmental and genetic risk factors contribute to the disease development. Through cumulative evidence, MED12 somatic mutations are emerging as an important player and an underlying cause of >70% ULMs.3 These mutations disrupt the kinase activity of the mediator complex and result in the dysregulation of the genes that are transcribed by RNA POLII, including, protein coding genes and functional RNAs.4,5 Recently, the role of long non-coding RNAs (lncRNAs) has been confirmed in several human diseases; however, there are a few studies regarding the role of these functional RNAs in ULM. Guo et al showed for the first time that lncRNAs, specially intragenic lncRNAs, are dysregulated in the uterine leiomyomas and concluded that the abnormal expression of lncRNAs may contribute to the pathogenesis of the disease.6 LncRNA SRA1 (SRA) is an intragenic lncRNA transcribed from the steroid receptor RNA activator1 (SRA1) gene located at 5q31.3. SRA RNA is capable to coactivate several nuclear receptors including, estrogen receptors (ERs).7–9 It is well understood that ULMs are estrogen-dependent tumors and estrogen exerts its tumorigenesis effect on ULM through binding to estrogen receptors , ERα and ERβ. Therefore, lncRNA SRA1 serves as a good target for investigating the possible mechanisms involved in the genesis of ULMs. The aim of the present study was to investigate the expression of lncRNA SRA1 in ULM samples regarding the mutation profile of MED12 gene.
Materials and methods
Patients and tissue samples
Sixty leiomyoma tissues were obtained from 49 Iranian women who were diagnosed with uterine leiomyoma and underwent an abdominal hysterectomy or myomectomy in Moheb-e-Yas hospital (Tehran, Iran). All samples were immediately frozen in liquid nitrogen and stored at −80°C until used. All participants provided written informed consent prior to study enrolment. This study was conducted in accordance with the ethical principles of the World Medical Association’s Declaration of Helsinki and was approved by the ethics committee of the Shahid Beheshti University of Medical Sciences (SBMU) (Code: IR.SBMU.MSP.REC.1398.261).
Mutation analysis
Genomic DNA was extracted from 30 mg of the tissues using DNeasy Blood & Tissue Kit (Qiagen, Cat No: 69504) according to the manufacturer protocol. To analyze mutations in MED12 exon 1, exon 2 and the flanking intronic regions, genomic DNA was amplified by PCR using two sets of specific primers as shown in Table 1. PCR reactions were prepared separately for each set of primers in a total volume of 25 µL, containing 12.5 µL Taq DNA Polymerase 2X Master Mix Red (Amplicon, Odense, Denmark), 1 µL of each primer (10 pmol), 2 µL genomic DNA (≥100 ng), and 8.5 µL PCR grade water. PCR was performed in a GeneTouch thermocycler instrument (BIOER, Hangzhou, China) with the following program: initial denaturation at 94°C for 5 min and a subsequent series of 35 cycles of 94°C for 30 s (denaturation), 61°C for 45 s (annealing), and 72°C for 30 s (extension). The final elongation step was performed at 72°C for 5 min. To define genomic alterations in MED12 gene, Sanger sequencing was performed using an ABI 3730xl DNA analyzer (Macrogen, Seoul, Korea). MED12 NG_012808.1 and NM_005120.2 were used as genomic and cDNA reference sequences, respectively. The A of the first ATG codon was considered as +1 in nucleotide numbering. Mutation analysis was performed using Chromas software (version:2.13).
 |
Table 1 Primer sequences for mutation detection and qRT-PCR |
Expression analysis
Total RNA was extracted from leiomyoma samples and adjacent tissues using easy-BLUE™ Total RNA Extraction Kit (iNtRON Biotechnology, Inc., Cat No: 17061). The first-strand cDNA was synthesized from 2 μg of total RNA by using High-Capacity cDNA Reverse Transcription Kit, according to the manufacturer-provided protocol (Applied Biosystems, Cat No: 4368813).
Quantitative RT-PCR (qRT-PCR) was performed for SRA1 lncRNA and beta-2-microglobulin (β2M) genes in duplicate on a Rotor Gene 6000. Each PCR reaction comprised of 10 µl 2X RealQ-PCR Master Mix® (Ampliqon, Odense, Denmark), 1 µl cDNA (≥10 ng), 0.5 µl of each primer (5 pmol) and 7 µl of nuclease-free water in a total volume of 20 µl reaction mixture. Primers for amplification of SRA1 lncRNA were obtained from a previous study.8
Three-step PCR thermal cycling and real-time data acquisition were performed using the following conditions: 95°C for 15ʹ ×1 cycle, and 95°C for 15 s, followed by 60°C for 15” and 72°C for 15” ×40 cycles followed by the melt curve stage assessment. To check the primer specificity, melting curves analysis and agarose gel (2%) electrophoresis were performed. The relative expression level of SRA1 lncRNA was normalized to the expression level of β2M.
Statistical analysis
Prior to the expression analysis, the cycle threshold (Ct) data and amplification efficiency for each reaction were calculated using LinRegPCR software (version: 2017.1). The gene expression ratio (fold change) for the lncRNA SRA1 in MED12 mutant and MED12 wild-type leiomyoma samples was calculated using the REST© 2009 software (v2.0.13). Unpaired-two-tailed Student's t-test was used to evaluate differences in the tumor size and gene expression level between the studied groups by using GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA). P<0.05 was considered statistically significant.
Results
The patients’ ages ranged from 29 to 54 years (42.87±7.75 years). The leiomyoma tissues were from three different types including intramural, subserousal, and submucousal with the frequency of 48.21%, 37.51%, and 14.28%, respectively. DNA sequencing analysis revealed somatic mutations in exon 2 of the MED12 gene in 28 (46.67%) ULM samples; including, 24 (85.71%) missense mutations and 4 (14.29%) in-frame deletions (Table 2 and Figure 1). No mutation was detected in the MED12 exon 1.
 |
Table 2 Detected MED12 mutations in the studied uterine leiomyomas |
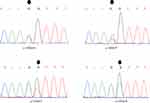 |
Figure 1 Chromatograms presenting some of the exon2-MED12 somatic mutations. |
The tumor size was statistically different between ULM samples with (2.32±0.40) and without (4.65±0.66) MED12 mutation (P-value=0.0356) (Figure 2).
 |
Figure 2 Comparing the tumor size between MED12-mutated and MED12-wild type leiomyoma tissues. *P≤0.05. |
We analyzed the expression level of lncRNA SRA1 in the uterine leiomyoma samples with and without mutation. LncRNA SRA1 was over-expressed in the ULM samples without MED12 mutation compared with ULM samples harboring MED12 mutation (Expression ratio =2.5, P-value=0.004). Figure 3 shows the compared delta Ct values between the studied groups.
Discussion
In the present study, we investigated the expression level of lncRNA SRA1 in ULM samples with and without MED12 mutation. Our results showed that the expression of the lncRNA SRA1 was up-regulated in the ULM samples negative for MED12 mutations in compared with ULM samples harboring these mutations (expression ratio =2.5; P-value=0.004). We also found that there was a negative correlation between the tumor size and MED12 mutation. Therefore, it may be concluded that up-regulation of lncRNA SRA1 affects the tumor size, indirectly.
As it was mentioned, SRA1 is a nuclear receptor that encodes both non-coding (lncRNA SRA1) and coding (SRAP) products result from alternative splicing.7,8,10 It is believed that both transcripts are functional and regulate several molecular pathways especially through ERs.7,9,11 SRA functions as a scaffold and several molecules including co-activators of nuclear receptors and transcription factors bind to it either directly or indirectly, and therefore regulate the gene expression.12 SRA co-activates both ERα and ERβ in a ligand-independent and ligand-dependent manner, respectively.12 The effect of SRA on the expression of ERα and ERβ is interesting, since both receptors play role in developing ULMS.13–16
According to the recent studies, both estrogen and progesterone hormones are important in developing ULMs. ERα and ERβ mediate the action of estrogens in the formation and growth of the leiomyoma; however, it is believed that the most estrogen action in the uterine attributes to ERα. In vitro studies suggest that the β form also modulates the ERα effects in the uterus.13
Previous studies have revealed that the size of ULM is different regarding the MED12 mutation pattern of the leiomyoma. It is suggested that ULMs harboring mutations in the MED12 exon 2 to be smaller in size compared with ULMs negative for these mutations.17–19 According to the present results, lncRNA SRA1 may explain this difference in the tumor size due to its role in the regulation of ERs.
Although the main fractions of the ULMs are smooth muscle cells; however, there are also several other components, including vessels, fibroblasts, and mast cells. The distribution of ERα and ERβ is quite different in ULM components. While ERα is found mostly in the uterine smooth muscle, ERβ is located in smooth muscle cells of the uterine, blood vessels endothelial cells, endothelial cells, and mast cells. The wide distribution pattern of ERβ suggests that it may regulate the expression of several factors necessary for tumor proliferation including, fibroblast growth factor, vascular endothelial growth factor (VEGF), and molecules of the extracellular matrix. In addition, due to its coexistence with ERα in uterine smooth muscle, ERβ may modulate the function of ERα in the uterine.13 Bakas et al showed that ERβ is over-expressed in ULM tissue compared with the normal uterine tissue.16
Valladares et al suggested that the expression of the ERβ in the ULM connective tissue cells may explain the abundant presence of extracellular matrix that is one of the structural characteristics of ULMs. All these evidences suggest that ERβ is consistent with growth favor in leiomyoma.
Moreover, comparing multiple and solitary leiomyomas revealed that ERβ level was significantly higher in the former, while the ERα was significantly lower in the same type of leiomyoma. Although in both types of leiomyoma, the ERα expression level was greater than the expression level of ERβ.20
The same as leiomyoma, endometriosis is an estrogen-dependent disease. According to Han et al, ERβ inhibits the cell death induced by TNFα and increases the proliferative activities of endometriotic tissues.21 Lin et al showed that siRNA-induced inhibition of SRA1 results in ESCs growth reduction due to the enhancement of the apoptosis and proliferation reduction of ESCs in endometriosis.9 It seems that SRA1 blocks the hypomethylation of the ERβ promoter region; therefore, SRA1 siRNA treatment causes ERβ down-regulation that itself results in the ERα up-regulation.
The main limitation of the present study is the small sample size since we classified the studied groups due to the MED12 mutation pattern. To our knowledge, this is the first study that indicates a functional role for lncRNA SRA1 in bound up with MED12 somatic mutation in uterine leiomyoma.
In conclusion, fibroids are estrogen-dependent and, understanding the factors that influence the expression of ERs is important for increasing our knowledge about the disease pathobiology and development of new therapeutics. Hence, lncRNA SRA1may be a good target, and provide a new vision into understanding the disease molecular mechanism.
Acknowledgment
The present article is financially supported by “Research Department of the School of Medicine Shahid Beheshti University of Medical Sciences” (Code: 17314).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Stewart EA, Cookson C, Gandolfo RA, Schulze‐Rath R. Epidemiology of uterine fibroids: a systematic review. BJOG. 2017. doi:10.1111/1471-0528.14640
2. Donnez J, Dolmans -M-M. Uterine fibroid management: from the present to the future. Hum Reprod Update. 2016;22(6):665–686. doi:10.1093/humupd/dmw023
3. Croce S, Chibon F. MED12 and uterine smooth muscle oncogenesis: state of the art and perspectives. Eur J Cancer. 2015;51(12):1603–1610. doi:10.1016/j.ejca.2015.04.023
4. Petrenko N, Jin Y, Wong KH, Struhl K. Mediator undergoes a compositional change during transcriptional activation. Mol Cell. 2016;64(3):443–454. doi:10.1016/j.molcel.2016.09.015
5. Jeronimo C, Langelier M-F, Bataille AR, Pascal JM, Pugh BF, Robert F. Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol Cell. 2016;64(3):455–466. doi:10.1016/j.molcel.2016.09.002
6. Guo H, Zhang X, Dong R, et al. Integrated analysis of long noncoding RNAs and mRNAs reveals their potential roles in the pathogenesis of uterine leiomyomas. Oncotarget. 2014;5(18):8625. doi:10.18632/oncotarget.2349
7. Cooper C, Guo J, Yan Y, et al. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009;37(13):4518–4531. doi:10.1093/nar/gkp441
8. Yan Y, Cooper C, Hamedani MK, et al. The steroid receptor RNA activator protein (SRAP) controls cancer cell migration/motility. FEBS Lett. 2015;589(24Pt B):4010–4018. doi:10.1016/j.febslet.2015.11.007
9. Lin K, Zhan H, Ma J, et al. Silencing of SRA1 regulates ER expression and attenuates the growth of stromal cells in ovarian endometriosis. Reprod Sci. 2017;24(6):836–843. doi:10.1177/1933719116670036
10. Hube F, Guo J, Chooniedass-Kothari S, et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25(7):418–428. doi:10.1089/dna.2006.25.418
11. Chooniedass-Kothari S, Vincett D, Yan Y, et al. The protein encoded by the functional steroid receptor RNA activator is a new modulator of ER alpha transcriptional activity. FEBS Lett. 2010;584(6):1174–1180. doi:10.1016/j.febslet.2010.02.024
12. Colley SM, Leedman PJ. Steroid receptor RNA activator - A nuclear receptor coregulator with multiple partners: insights and challenges. Biochimie. 2011;93(11):1966–1972. doi:10.1016/j.biochi.2011.07.004
13. Valladares F, Frias I, Baez D, et al. Characterization of estrogen receptors alpha and beta in uterine leiomyoma cells. Fertil Steril. 2006;86(6):1736–1743. doi:10.1016/j.fertnstert.2006.05.047
14. Borahay MA, Asoglu MR, Mas A, Adam S, Kilic GS, Al-Hendy A. Estrogen receptors and signaling in fibroids: role in pathobiology and therapeutic implications. Reprod Sci. 2017;24(9):1235–1244. doi:10.1177/1933719116678686
15. Burns KA, Korach KS. Estrogen receptors and human disease: an update. Arch Toxicol. 2012;86(10):1491–1504. doi:10.1007/s00204-012-0868-5
16. Bakas P, Liapis A, Vlahopoulos S, et al. Estrogen receptor alpha and beta in uterine fibroids: a basis for altered estrogen responsiveness. Fertil Steril. 2008;90(5):1878–1885. doi:10.1016/j.fertnstert.2007.09.019
17. Markowski DN, Bartnitzke S, Löning T, Drieschner N, Helmke BM, Bullerdiek J. MED12 mutations in uterine fibroids—their relationship to cytogenetic subgroups. Int J Cancer. 2012;131(7):1528–1536. doi:10.1002/ijc.27424
18. Heinonen H-R, Sarvilinna NS, Sjöberg J, et al. MED12 mutation frequency in unselected sporadic uterine leiomyomas. Fertil Steril. 2014;102(4):1137–1142. doi:10.1016/j.fertnstert.2014.06.040
19. Mäkinen N, Mehine M, Tolvanen J, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334(6053):252–255. doi:10.1126/science.1208930
20. Shao R, Fang L, Xing R, Xiong Y, Fang L, Wang Z. Differential expression of estrogen receptor alpha and beta isoforms in multiple and solitary leiomyomas. Biochem Biophys Res Commun. 2015;468(1–2):136–142. doi:10.1016/j.bbrc.2015.10.145
21. Han SJ, Jung SY, Wu S-P, et al. Estrogen receptor β modulates apoptosis complexes and the inflammasome to drive the pathogenesis of endometriosis. Cell. 2015;163(4):960–974. doi:10.1016/j.cell.2015.10.034
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

