Back to Journals » Cancer Management and Research » Volume 12
LncRNA SNHG9 is Downregulated in Non-Small Cell Lung Cancer and Suppressed miR-21 Through Methylation to Promote Cell Proliferation
Authors Wang D, Cao X, Han Y, Yu D
Received 9 March 2020
Accepted for publication 25 June 2020
Published 31 August 2020 Volume 2020:12 Pages 7941—7948
DOI https://doi.org/10.2147/CMAR.S253052
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Dingxue Wang,1,* Xiaoqing Cao,2,* Yi Han,2 Daping Yu2
1Department of Oncology, The First Affiliated Hospital of Guizhou University of Traditional Chinese Medicine, Guiyang, 550001, People’s Republic of China; 2Department of Thoracic Surgery, Beijing Chest Hospital, Capital Medical University, Beijing Tuberculosis and Thoracic Tumor Research Institute, Beijing 101149, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Daping YuDepartment of Thoracic Surgery
Beijing Chest Hospital, Capital Medical University, Beijing Tuberculosis and Thoracic Tumor Research Institute, Area 1st, No. 9 Compound, Beiguan Street, Tongzhou District, Beijing 101149, People’s Republic of China
Email [email protected]
Background: LncRNA SNHG9 has been shown to be an oncogenic lncRNA in glioblastoma, while its role in other cancers is unknown. The aim of this study was to investigate the role of SNHG9 in non-small cell lung cancer (NSCLC).
Methods: The differential expression of SNHG9 in NSCLC was first explored by analyzing the TCGA dataset, followed by measuring the expression levels of SNHG9 in paired NSCLC and non-tumor tissues by RT-qPCR. Expression of miR-21 was also determined by RT-qPCR. Correlations were analyzed by linear regression. The interaction between miR-21 and SNHG9 was detected using RNA pull-down. The expression relationship between SNHG9 and miR-21 was analyzed by SNHG9 or miR-21 overexpression experiments. The effects of overexpression of SNHG9 on the methylation of miR-21 were analyzed by methylation-specific PCR (MSP). Cell proliferation was evaluated by CCK-8 assay.
Results: By analyzing the TCGA dataset, we observed downregulation of SNHG9 in NSCLC, which was confirmed by measuring the expression levels of SNHG9 in paired NSCLC tumor tissues and non-tumor tissues from NSCLC patients involved in this study. MiR-21 was upregulated in NSCLC tumor tissues and inversely correlated with SNHG9 in cancer tissues but not in non-tumor tissues. The interaction between SNHG9 and miR-21 was predicted by bioinformatic analyses, which was further verified by RNA pull-down. In NSCLC cells, overexpression of SNHG9 led to downregulated miR-21 and increased methylation of miR-21 gene. In contrast, miR-21 did not affect the expression of SNHG9. In addition, overexpression of SNHG9 attenuated the enhancing effects of miR-21 on NSCLC proliferation.
Conclusion: SNHG9 might downregulate miR-21 through methylation to suppress cancer cell proliferation.
Keywords: SNHG9, non-small cell lung cancer, miR-21, methylation
Introduction
Lung cancer is the most common type of malignancy and the major cause of cancer-related deaths in both males and females.1 It has been reported that lung cancer in 2018 alone affected a total number of 2,093,876 new cases, accounting for 11.6% of all cancer cases, and caused 1,761,007 deaths, accounting for 18.4% of cancer deaths.2 Most cases of lung cancer are diagnosed at advanced stages with tumor spread already.3,4 With tumor metastasis, only less than 5% of lung cancer patients can live for more than 5 years after the initial diagnosis. Although smoking has been proven to be the major cause of lung cancer,5,6 never-smokers are also affected by this disease,7 indicating its complicated pathogenesis.
Studies on the molecular pathogenesis of lung cancer increased the understanding of the mechanisms underlying the occurrence, development and progression of lung cancer.8,9 A considerable number of molecular players have been revealed to promote or inhibit the progression of lung cancer.8,9 In fact, these factors are potential targets for the development of novel anti-cancer therapies, such as targeted therapy.10,11 Although non-coding RNAs (ncRNAs), such as long (>200 nt) ncRNAs (lncRNAs) and miRNAs, are not involved in protein-coding, but they regulate gene expression to participate in cancer biology.12 Therefore, ncRNAs are novel targets for cancer treatment. However, the functionality of most ncRNAs remains unclear. LncRNA SNHG9 has been shown to be involved in several types of cancer, such as glioblastoma and pancreatic cancer (PC).13,14 It has been reported that the expression of SNHG9 was upregulated in glioblastoma tissues and its increase was related to lower survival rate in patients with glioma. And overexpression of SNHG9 in glioblastoma cells promoted aerobic glycolysis and cell proliferation through regulating the miR-199a-5p/Wnt2 axis.13 Another research focused on the regulatory roles of lncRNA SNHG9 in the pathogenesis of PC demonstrated that SNHG9 were downregulated in PC tissues and serums, and their expression levels were correlated with clinical stages and survival rate of PC patients.14 All these studies have provided the evidence for the important roles of SNHG9 in the progression of different cancers.
The effects of miR-21 have been widely investigated in different diseases including lung cancer.15 MiR-21 was one of the first miRNAs identified in mammalian, which is located on chromosome 17 (17q.23.1) in the 11th intron of the TMEM49 (transmembrane protein 49) gene, precursor of VMP1 (vacuole membrane protein 1).16–18 It has been revealed that the upregulation of miR-21 promoted cell migration and cell growth in human non-small cell lung cancer cells through CD36 mediated fatty acid metabolism.15 In addition, it was reported that the expression levels of miR-21 were elevated in NSCLC, and the expression of miR-21 was inversely correlated with overall survival of NSCLC patients, which indicated that miR-21 promoted the development of NSCLC.19 In our study, we analyzed the TCGA dataset and observed downregulation of SNHG9 and its inverse correlation with miR-21 as well as its inhibitory roles in the regulation of the proliferation of non-small cell lung cancer (NSCLC) cells.
Materials and Methods
NSCLC Patients
A total of 64 NSCLC patients (48 males and 16 females, age range 45 to 70 years old, mean age 57.4 ± 5.0 years old) were enrolled at this hospital between May 2017 and October 2019. The diagnosis of NSCLC was performed by imaging and histopathological biopsy. Patients with other severe clinical disorders were excluded. All patients have no family history or previous history of malignancies. Based on AJCC staging and clinical findings, the 64 patients were divided into stage I (n = 12), II (n = 10), III (n = 21) and IV (n = 21).
Patient Pre-Treatment
In total, 64 paired NSCLC cancer and non-tumorous tissues samples were obtained from patients undergoing a surgical procedure at the Beijing Chest Hospital. All patients had signed the written informed consent, in which the key information including voluntary participation, collection and use of patients’ samples only for scientific research, publication of their individual-level data, confidentiality guarantee, detailed design of the study and no risk or influence on their following clinical diagnosis and treatment was clearly described. And all experiments were performed in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki). This study (including the collection and use of patients’ samples) was approved by the Ethics Committee of the Beijing Chest Hospital.
NSCLC Tissues and Cells
Paired NSCLC and non-tumor lung tissues were obtained from each patient through biopsy. All tissue samples were confirmed by histopathological analysis and were stored in a liquid nitrogen tank before use. Two NSCLC cell lines, H1581 and H1993, were purchased from ATCC (USA). Cells were cultivated in a 5% CO2 incubator at 37 °C. Cell culture medium was composed of RPMI-1640 medium (90%) and FBS (10%). SNHG9 expression vector was constructed using pcDNA3.1 backbone vector (Invitrogen). Mimic of miR-21 and control miRNA were provided by Sangon (Shanghai, China). H1581 and H1993 cells were harvested at about 85% confluence, followed by the transfection of cells with either vector (10 nM) or miRNA (40 nM) using Lipofectamine 2000 (Invitrogen). PcDNA3.1 empty vector or control miRNA-transfected cells were annotated as negative control (NC) group. Untransfected cells were used as control (C). Subsequent analyses were performed at 48 h post-transfection. Lipofectamine 2000 (Invitrogen) was used as transfection reagent.
Bioinformatic Analyses
The interaction between SNHG9 and miR-21 was predicted using an online program IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp). During prediction, miR-21 sequence was used as the “short sequence” and SNHG9 sequence was used as the “long sequence”.
RNA Pull-Down Assay
For miRNA pull-down, H1581 and H1993 cells were transfected with biotinylated miR-21 (miR-21 probe) or control probe (Genescript, Nanjing, China) and harvested in lysis buffer that contains 20 mM Tris pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.5% NP-40 and 1 U/ul Recombinant RNAse inhibitor (TaKaRa). Total RNAs samples were pretreated with DNaseI and then heated at 65 °C for 5 min, followed by an instant ice bath. RNA samples were then incubated with streptavidin-coated magnetic beads (New England BioLabs, S1420S) at 4 °C for 4 h. After incubation, beads were washed twice with lysis buffer and RNAs were extracted with Trizol (Invitrogen, CA, USA), and the expression of lncRNA SNHG9 was detected by RT-qPCR.
RNA Preparation
Total RNAs were extracted from paired tissues, H1581 and H1993 cells using TRIzol (Sigma-Aldrich) reagent. Genomic DNA removal was performed using DNase I. A total of 3 µg RNA sample was used for subsequent RT-qPCR.
RT-qPCR
SRT IV system (Thermo Fisher Scientific) was used to carry out reverse transcriptions (RTs), following by qPCRs performed using SYBR Green Master Mix (Bio-Rad) to determine the expression of SNHG9. 18S rRNA was used as the internal control for SNHG9. Ploy (A) was added to mature miRNAs using All-in-One™ miRNA qRT-PCR Reagent Kit (Genecopoeia). The same kit was used to perform miRNA RTs and qPCRs to measure the expression levels of mature miR-21. U6 was used as the internal control for miR-21. Triplicate reactions were included in each experiment. Data were normalized using 2−ΔΔCT. The primers sequences were as follows:
SNHG9 forward: 5ʹ-GACTGCAGACCCCTAACCTT-3ʹ,
SNHG9 reverse: 5ʹ-ACCCGCATGCAGTGAGTTA-3ʹ,
miR-21 forward: 5ʹ-GGACTAGCTTATCAGACTG-3ʹ,
miR-21 reverse: 5ʹ-GGACTAGCTTATCAGACTG-3ʹ,
U6 forward: 5ʹ-GC TTCGGCAGCACATATACTAAAAT-3ʹ,
U6 reverse: 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3′.
Methylation-Specific PCR (MSP)
Genomic DNAs were extracted from H1581 and H1993 cells using Monarch® Genomic DNA Purification Kit (NEB). DNA Methylation-GoldTM kit (ZYMO RESEARCH) was used to convert all DNA samples. With converted DNA samples as template, Taq 2X master mix (NEB) was used to perform MSPs to measure the level of miR-21 gene methylation. After amplification, approximately 200 bp product was obtained for subsequent analysis. The primers sequences were as follows:
Methylation forward: TGTAAAACGACGGCCAGT,
Methylation reverse: CAGGAAACAGCTATGACC,
Unmethylation forward: TGTAAAACGACGGCCAGT,
Unmethylation reverse: CAGGAAACAGCTATGACC.
Cell Proliferation Analysis
Cell proliferation assay was performed using Cell Counting Kit-8 (CCK-8) kit (Dojindo) to analyze the proliferations of H1581 and H1993 cells after transfections. Cells were first subjected to 0.25% trypsin digestion, following by cell culture in a 96-well plate with 5000 cells in 0.1 mL medium per well. OD values of the medium were measured every 24 h for a total of 4 d. CCK-8 was added up to 10% before OD value measurement.
Statistical Analysis
Experiments were performed in triplicate and mean ± SD values were calculated. Paired t test was used to compare differences between paired tissue samples. ANOVA Tukey’s test was used to compare differences among multiple groups. Correlations were analyzed by Pearson’s correlation coefficient. P < 0.05 was statistically significant.
Results
SNHG9 Was Downregulated but miR-21 Was Upregulated in NSCLC
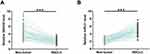
By analyzing the TCGA dataset, we observed downregulation of SNHG9 in both lung squamous cell carcinoma (LUSC; 24.9 vs 30.79) and lung adenocarcinoma (LUAD; 26.61 vs 30.21). To further confirm the downregulation of SNHG9 in NSCLC, the expression of SNHG9 in paired NSCLC tumor tissues and non-tumor tissues from 64 NSCLC patients was analyzed by performing RT-qPCR. It was observed that expression levels of SNHG9 were significantly lower in NSCLC tumor tissues compared to that in non-tumor tissues (Figure 1A, p < 0.001). The expression of miR-21 in paired NSCLC and non-tumor tissues from NSCLC patients were also analyzed by performing RT-qPCR. Compared to non-tumor tissues, the expression levels of miR-21 were significantly higher in NSCLC tissues (Figure 1B, p < 0.001).
SNHG9 and miR-21 Were Inversely Correlated in NSCLC and Interacted with Each Other
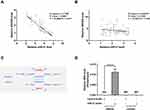
In order to investigate the correlation between SNHG9 and miR-21, the expression levels of them were correlated by detecting RNAs extracted from paired tumor tissues and non-tumor tissues, respectively. Correlation analysis showed that the expression levels of miR-21 and SNHG9 were inversely and significantly correlated with each other across NSCLC tissues (Figure 2A), but not in non-tumor tissues (Figure 2B). The interaction between SNHG9 and miR-21 was assessed by an online program IntaRNA, which showed that there was a binding site with miR-21 at 3ʹ-UTR of the SNHG9 gene (Figure 2C). RNA pull-down assay was conducted to detect the binding between them. LncRNA SNHG9 could only be precipitated by miR-21 probe but it was undetectable in the product precipitated by scramble control probe (Figure 2D), indicating that miR-21 directly interacts with SNHG9.
Overexpression of SNHG9 Promotes the Methylation of miR-21
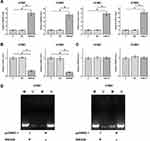
H1581 and H1993 cells were transfected with either SNHG9 expression vector or miR-21 mimic to explore the expression relationship between SNHG9 and miR-21. Overexpression of SNHG9 and miR-21 was confirmed by RT-qPCR (Figure 3A, p < 0.05). It was observed that overexpression of SNHG9 downregulated miR-21 (Figure 3B, p < 0.05), while overexpression of miR-21 did not affect the expression of SNHG9 (Figure 3C). To explore the mechanism, MSP was performed to analyze the effect of overexpression of SNHG9 on the methylation of miR-21 gene. Compared to cells transfected with empty pcDNA3.1 vector, cells transfected with SNHG9 expression vector showed increased methylation of miR-21 gene (Figure 3D).
SNHG9 Suppressed NSCLC Cell Proliferation Through Modulating miR-21
The role of SNHG9 and miR-21 in regulating NSCLC cell proliferation was analyzed by performing cell proliferation assay. It was observed that overexpression of SNHG9 resulted in decreased cell proliferation rate, and overexpression of miR-21 led to increased cell proliferation rate. Moreover, overexpression of SNHG9 attenuated the enhancing effects of miR-21 on NSCLC proliferation (Figure 4, p < 0.05).
Discussion
The interactions between SNHG9 and miR-21 in lung cancer were explored in this study. We found that SNHG9 was downregulated in NSCLC and it suppressed the expression of miR-21 through methylation and then inhibited NSCLC cell proliferation.
The involvement of SNHG9 in cancer biology has been investigated in several types of malignancy.13 It has been reported that SNHG9 was upregulated in glioblastoma and could interact with the iR-199a-5p/Wnt2 axis to promote aerobic glycolysis and cell growth.13 Another study reported that SNHG9 was downregulated in prostate cancer and predicted the survival of patients.15 Therefore, SNHG9 may exhibit different expression patterns and play different roles in different types of malignancy. In our study, we found that SNHG9 was downregulated in NSCLC and its suppressive roles in NSCLC cell proliferation, which suggests that lncRNA SNHG9 may participate in the progression of NSCLC.
MiR-21 is a well-studied oncogenic miRNA in cancer biology.20 MiR-21 targets several tumor suppressors, such as PTEN and programmed cell death to inhibit cancer development and progression.19,21 However, the upstream regulators of miR-21 in cancer biology still have not been precisely explored. In this study, we first observed the upregulation of miR-21 in NSCLC tumor tissues relative to non-tumor tissues and its inverse correlation with lncRNA SNHG9 in NSCLC, which suggests SNHG9 and miR-21 have opposite expression patterns in NSCLC, and their correlation might be an important internal mechanism in the regulation of NSCLC. In order to verify whether there is an interaction between SNHG9 and miR-21, we conducted bioinformatic analyses and RNA pull-down assay. Bioinformatic analyses showed that a binding site with miR-21 was located at the 3ʹ-UTR of SNHG9 gene, and lncSNHG9 existed in the immunoprecipitated pellet by miR-21 probe but not scramble control probe. To further investigate the regulatory relationship between SNHG9 and miR-21, SNHG9 or miR-21 was overexpressed in NSCLC cell lines. Interestingly, overexpression of SNHG9 significantly suppressed the expression of miR-21. However, miR-21 had no responsive effect on SNHG9. Our MSP experiment reveals that overexpression of SNHG9 could promote the methylation of miR-21 gene in NSCLC cell lines, suggesting the mastery of SNHG9 to miR-21 might be taken through gene methylation. MiR-21 can impact cancer cell proliferation via targeting different signaling proteins.22,23 In our study, we also found that miR-21 promoted the proliferation of NSCLC cells, which was consistent with previous studies that miR-21 deteriorates the progression of NSCLC.19 As expected, SNHG9 had suppressive effect on NSCLC cell proliferation, indicating its protective role in NSCLC progression.
It is worth noting that the expression levels of SNHG9 and miR-21 were inversely and significantly correlated only across NSCLC tissues but not non-tumor tissues. Thus, the interaction between SNHG9 and miR-21 may only occur under pathological factors. One possible hypothesis is that some methylation factors were altered in NSCLC cells, and the altered methylation factors mediate the interaction between SNHG9 and miR-21. However, future studies are needed to perform deeper investigations.
In conclusion, SNHG9 is inhibited in NSCLC and miR-21 is upregulated in NSCLC. SNHG9 may downregulate miR-21 through methylation to suppress NSCLC cell proliferation.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Alberg AJ, Samet JM. Epidemiology of lung cancer. Chest. 2003;123(1):21S–49S. doi:10.1378/chest.123.1_suppl.21S
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
3. Adjei AA. Lung cancer worldwide. J Thorac Oncol. 2019;14(6):956. doi:10.1016/j.jtho.2019.04.001
4. Hoffman RM, Sanchez R. Lung cancer screening. Med Clin North Am. 2017;101(4):769–785. doi:10.1016/j.mcna.2017.03.008
5. O’Keeffe LM, Taylor G, Huxley RR, et al. Smoking as a risk factor for lung cancer in women and men: a systematic review and meta-analysis. BMJ Open. 2018;8(10):e021611. doi:10.1136/bmjopen-2018-021611
6. Pierce JP, Shi Y, McMenamin SB, et al. Trends in lung cancer and cigarette smoking: california compared to the rest of the United States. Cancer Prev Res (Phila). 2019;12(1):3–12. doi:10.1158/1940-6207.CAPR-18-0341
7. Hung RJ, Spitz MR, Houlston RS, et al. Lung cancer risk in never-smokers of European descent is associated with genetic variation in the 5p15. 33 TERT-CLPTM1Ll region. J Thorac Oncol. 2019;14(8):1360–1369. doi:10.1016/j.jtho.2019.04.008
8. Kadara H, Scheet P, Wistuba II, et al. Early events in the molecular pathogenesis of lung cancer. Cancer Prev Res (Phila). 2016;9(7):518–527. doi:10.1158/1940-6207.CAPR-15-0400
9. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi:10.1038/nature13385
10. Chan BA, Hughes BGM. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015;4(1):36–54.
11. Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer—is it becoming a reality? Nat Rev Clin Oncol. 2010;7(7):401–414. doi:10.1038/nrclinonc.2010.64
12. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45(8):1895–1910. doi:10.1016/j.biocel.2013.05.030
13. Zhang H, Qin D, Jiang Z, et al. SNHG9/miR-199a-5p/Wnt2 axis regulates cell growth and aerobic glycolysis in glioblastoma. J Neuropathol Exp Neurol. 2019;78(10):939–948. doi:10.1093/jnen/nlz078
14. Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res. 2018;10(8):2648–2658.
15. Ni K, Wang D, Xu H, et al. miR-21 promotes non-small cell lung cancer cells growth by regulating fatty acid metabolism. Cancer Cell Int. 2019;19:219. doi:10.1186/s12935-019-0941-8
16. Cai X, Hagedorn CH, Cullen BR, et al. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2014;10(12):1957–1966. doi:10.1261/rna.7135204
17. Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi:10.1126/science.1064921
18. Ribas J, Ni X, Castanares M, et al. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res. 2012;40(14):6821–6833. doi:10.1093/nar/gks308
19. Xue X, Liu Y, Wang Y, et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget. 2016;7(51):84508–84519. doi:10.18632/oncotarget.13022
20. Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl). 2016;7:19–27. doi:10.2147/LCTT.S60341
21. Zhang J, Wang J, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta. 2010;411(11–12):846–852. doi:10.1016/j.cca.2010.02.074
22. Zhao Q, Chen S, Zhu Z, et al. miR-21 promotes EGF-induced pancreatic cancer cell proliferation by targeting Spry2. Cell Death Dis. 2018;9(12):1157. doi:10.1038/s41419-018-1182-9
23. Du X, Hong L, Sun L, et al. miR-21 induces endothelial progenitor cells proliferation and angiogenesis via targeting FASLG and is a potential prognostic marker in deep venous thrombosis. J Transl Med. 2019;17(1):270. doi:10.1186/s12967-019-2015-z
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.