Back to Journals » Cancer Management and Research » Volume 13
LncRNA SNHG8 Serves as an Oncogene in Breast Cancer Through miR-634/ZBTB20 Axis
Authors Xu X, Xie Q, Xie M, Zeng Y, Liu Q
Received 5 August 2020
Accepted for publication 16 December 2020
Published 7 April 2021 Volume 2021:13 Pages 3017—3028
DOI https://doi.org/10.2147/CMAR.S270128
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Xianyun Xu,1,* Qiongjun Xie,2,* Mingfeng Xie,3 Yong Zeng,3 Qian Liu1
1Key Laboratory of Prevention and treatment of cardiovascular and cerebrovascular diseases of Ministry of Education, Jiangxi Provincial Clinical Research Center for Vascular Anomalies, Basic Medical School, Gannan Medical University, Ganzhou, Jiangxi, 341000, People’s Republic of China; 2Basic Medical School, Gannan Medical University, Ganzhou, Jiangxi, 341000, People’s Republic of China; 3Department of Pediatric Surgery, Jiangxi Provincial Clinical Research Center for Vascular Anomalies, Key Laboratory of Prevention and Treatment of Cardiovascular and Cerebrovascular Diseases of Ministry of Education, The First Affiliate Hospital of Gannan Medical University, Ganzhou, Jiangxi, 341000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qian Liu
Key Laboratory of Prevention and treatment of cardiovascular and cerebrovascular diseases of Ministry of Education, Jiangxi Provincial Clinical Research Center for Vascular Anomalies, Basic Medical School, Gannan Medical University, Ganzhou, Jiangxi, 341000, People’s Republic of China
Email [email protected]
Background: Small nucleolus RNA Host Gene 8 (SNHG8) belongs to a subgroup with long non-coding RNAs. LncRNA SNHG8 presents up-regulated in miscellaneous cancers, like gastric cancer, liver cancer, and esophageal squamous cell cancer. Nevertheless, the expression pattern and the pathological function of lncRNA SNHG8 in breast cancer remain obscure.
Methods: We examined the expression levels of lncRNA SNHG8 in the tissue samples and cell lines from breast cancer via RT-qPCR in the present study. The functions of lncRNA SNHG8 on the progression of breast cancer cell were examined by CCK-8, EdU, Transwell chamber assays, and flow cytometry analyses. The expression of proteins was assessed using Western blot assay.
Results: We found that proliferation, migration, and invasion of breast cancer cells were significantly inhibited due to knockdown of lncRNA SNHG8, while inducing apoptosis of these cells. Mechanistically, SNHG8 functioned as an inhibitor of miR-634 in tumor tissues.
Conclusion: LncRNA SNHG8 sponged the miR-634 to increase the expression level of ZBTB20, thus further aggravating the malignancy of breast cancer. Hence, the lncRNA SNHG8-miR-634-ZBTB20 axis may be a promising therapeutic target to treat breast cancers.
Keywords: small nucleolus RNA host gene 8, microRNA-634, breast cancer, ZBTB20
Introduction
Breast cancer is one of the most common malignant tumors in women worldwide.1 In the past few decades, breast cancer has become the most common cancer among women, especially in developed countries as well as the second cause of women cancer death after lung cancer.1–3 However, the specific pathogenesis of breast cancer remains unclear.4,5 Previous studies have shown that not only genetic and endocrine factors but also the improper expressions of Lnc/miRs and proteins are involved in the occurrence of breast cancer.6,7
Long non-coding RNAs (lncRNA) are a group of RNAs that consist of over 200 nucleotides and have no protein-coding function except for guiding RNAs in post-translational modifications.8–10 It is well known that lncRNAs regulate gene expression through a variety of mechanisms, including genomic interactions, protein content, miRNA competition, and chromatin modification.9 Recent reports prove that lncRNAs feature in diverse biological and pathological behaviors, especially in carcinogenesis and cancer progression.10 At present, a multitude of lncRNAs have been reported to express abnormally in breast cancer. For example, Lnc-UCA1, Lnc-Smad7, and Lnc-LSINCT5 are reported to over-express in various breast cancers and play oncogenes roles in the malignant phenotypes.11 LncRNA SNHG8 has been reported to be over-expressed in various types of cancer, indicating that lncRNA SNHG8 may participate in the progression of these cancers.12–14 However, it has not been clarified whether lncRNA SNHG8 is abnormally expressed in breast cancer.
In our study, we found that lncRNA SNHG8 was increased in breast cancer samples and cell lines. Knockdown of lncRNA SNHG8 inhibited the cell proliferation, migration/invasion, while triggering cell apoptosis of breast cancer cell lines. At the molecular level, lncRNA SNHG8 sponged miR-634 to induce the expression of an oncogene ZBTB20, which further triggered the activation of breast cancer progression. Therefore, we provided a novel molecular mechanism correlated with the pathology in breast cancer and offered a novel direction for treating breast cancers.
Materials and Methods
Collection of Tissue Specimens
All breast cancer tissues and paired peritoneal tissues (n = 30) were obtained from patients with breast cancer. All the protocols concerning the usage of patient samples were authorized by the Medical Ethics Committee of The first affiliate Hospital of Gannan Medical University. Informed consent was signed by all patients participating in the study. This research was conducted abiding by the World Medical Association Declaration of Helsinki.
Cell Lines and Culture Conditions
Human breast cancer cell lines (MCF7, Hs-578T, ZR-75-30, HCC1973) and human normal breast cell MCF-10A were purchased from the Chinese Academy of Sciences, Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cells were maintained in DMEM (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone, USA) and grown in humidified 37°C incubators supplied with 5% of CO2. The CO2 cell incubator purchased from Forma Scientific UK and FACS Calibri flow cytometer purchased from BD biosciences (USA). ABI7300 fluorescence quantitative PCR instrument was purchased from Applied Biosystems Inc.
RNA Extraction and RT-qPCR
TRIZOL reagent (ThermoFisher, USA) was adopted to refine total RNA in cells and tissues. Taqman probes (ThermoFisher, USA) were used to quantify miRNAs. To be brief, 1 µg of total RNA was transcribed to cDNA with the help of AMV reverse transcriptase (Takara, Japan) and an RT primer. The reaction conditions were 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. Real-time quantitative PCR was performed by a Taqman PCR kit on an Applied Biosystems 7300 sequence detection systems (Thermofisher, USA). The reactions were performed in a 96-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 1 min. U6 and GAPDH were used as the internal control. SNHG8 upstream primer: 5ʹ-CCCGAGAACCGT-CAGTTTGA-3ʹ, downstream primer: 5ʹ-ACACCCGTTTCCCCAACTAC-3ʹ. MiR‐634 upstream primer: 5′‐CAGTCTCAAACCAGCACC‐3′, downstream primer: 5′‐TATGGTTGTTCACGACTCCTTCAC‐3′. ZBTB20 upstream primer: 5ʹ -GCTTGGCTACAGCGACATC-3ʹ, downstream primer: 5ʹ-GAACACATCGCCCACGTTC-3ʹ. U6 upstream primer: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, downstream primer: 5ʹ- AACGCTTCACGAATTTGCGT-3ʹ. GAPDH upstream primer: 5ʹ-AATCCCATCACCATCTTC-3ʹ, downstream primer: 5ʹ- AGGCTGTTGTCATACTTC-3ʹ.
Western Blotting Analysis
Samples were rifted in RIPA lysis buffer plus PMSF with low temperature, and BCA assay kit (Santa Cruz, California, USA) examined total protein concentration. Prepared protein samples were separated in SDS-PAGE and transferred into 0.22 μm PVDF membranes and then were incubated with prepared antibodies. Finally, this membrane was visualized by enhanced chemiluminescence (ThermoFisher, MA, USA). The antibody against β-actin was purchased from CST (Beverly, Massachusetts, USA). Antibodies against Bax, Bcl-2, MMP-2, and MMP-9 were purchased from Abcam (MA, USA).
Scratch Assay
When cells reached 90% confluence, a single wound was created and phase-contrast images were digitally photographed immediately after 24-h incubation. The original opening distance of the wound was set as 100%. The opening distances after 24 h were detected in three areas which were randomly selected from per well, and the distance in three wells of each group was quantified and normalized by the original opening distance. The experiment was performed three times in triplicate, and the percentage of the migration rate was calculated by examining the length of cell migration and expressed as a percentage in comparison with the control group. Migration rates = (treatment group cell migration distance/control group migration distance) × 100%.
Cell Counting Kit-8 (CCK-8) Assay
Cells were collected and resuspended in the culture medium after 24-h transfection. In aggregate, 100 μL of cell suspension carrying 2×103 cells was injected into 96-well plates. Cells were incubated in the humidified incubator under the circumstance of 37°C and 5% CO2 in order to determine the cellular proliferation at specific time points: 0, 1, 2, and 3 days after inoculation. At each time point, 10 μL of the CCK-8 solution (Syngene, Nanjing, China) was appended into each well, followed by incubation at 37°C for extra 2 h. The absorbance of each well was measured at a 450 nm wavelength on a micro-plate reader (Thermofisher, USA).
EdU Assay
Cell lines were incubated with EdU (Syngene, Nanjing, China) and fixed with 1 mL 4% paraformaldehyde (Syngene, Nanjing, China) at room temperature for 30 min, then cell lines were washed with 3% BSA twice, 5 min each time, and permeated with 1 mL osmotic agent (0.5% Triton X-100) to permeate the cytoplasm and nuclear membrane. After 20 min of permeation, each cell line was washed with 3% BSA twice for 5 min each time to remove the osmotic agent. 0.5 mL of the KeyFluor488 (Syngene, Nanjing, China) click-it reaction mixture was added to each culture dish and incubated at room temperature, for 30 min in darkness. Click-it reaction mixture was discarded, and the reaction mixture was rinsed with 3% BSA twice, 5 min each time. The reaction liquid was removed, add antifluorescence quenching agent and observed with a fluorescence microscope (ThermoFisher, MA, USA).
Transwell Chamber Assay
The capability of cell migration was evaluated by Transwell chambers (8 μm diameters; Shanghai, China). Cultured for 48 h, the transformed cells were harvested, washed with PBS, and resuspended in DMEM without FBS. In aggregate, 100 μL of suspension with 5×104 cells was appended into the upper chambers. The transwell chambers were put into a 24-well plate teemed with 500 μL of DMEM containing 10% of FBS. Twenty-four hours later, non-migratory cells were gently eliminated. Meanwhile, the migratory cells were fixed with 100% methanol, stained with 0.5% crystal violet, washed with PBS, and imaged by an inverted microscope (Olympus, Tokyo, Japan). The migratory and invasive abilities were detected by counting the migratory and invading cells, respectively.
Luciferase Reporter Assay
The luciferase reporter plasmids were constructed by cloning the SNHG8-WT, SNHG8-Mut (mutant of functional miR-634 binding domain), as well as ZBTB20-3ʹ-UTR-WT and ZBTB20-3ʹ-UTR-Mut into the psiCHECK-2 vectors (Synthgene Biotech, Nanjing, China). The sequences that could be binding to miR-634 were partly mutated and inserted into the reporter plasmids in order to identify the binding specificity. Briefly, MCF7 and ZR-75-30 cells were seeded in 24 well plates until reaching 60% confluence. Each well was co-transfected with luciferase reporter plasmids (0.5 μg) and miRNA mimics (100 nM) using Lipofectamine 2000 (Thermofisher, USA) according to the manufacturer’s protocol. The luciferase activity was measured after 48-h transfection by using the Dual-Luciferase Reporter Assay (Promega, Shanghai, China) in accordance with the manufacturer’s instructions and normalized to Renilla signals.
Plasmid Construction
Knockdown of the lncRNA SNHG8 and ZBTB20 was accomplished by transfecting cells with targeting shRNAs. shRNAs targeting the lncRNA SNHG8, ZBTB20, and control were synthesized by Synthgene (China). All the cells were transferred with them using Lipofectamine 2000 (Thermofisher, USA) complying with the manufacturer’s instruction. All samples were collected, respectively, after 24-h and 48-h transfection.
Statistical Analyses
All experiments were repeated three times and the data were presented as the mean standard deviation by using SPSS 18.0 (SPSS, Inc.). A paired t-test was performed to compare the mean of the two groups, and then a one-way ANOVA and Bonferroni multiple comparison test was adopted to examine differences between the two or more groups. P < 0.05 was considered statistically indispensable.
Results
The Expression Level of SNHG8 is Up-Regulated Both in Breast Cancer Tissues and Its Cell Lines
Firstly, we measured its expression pattern in the 30 pairs of breast cancer and normal tissues as it seeks to evaluate the latent regulatory roles of SNHG8 in breast cancer. RT-qPCR analysis suggested that the expression level of SNHG8 was increased in tumor tissues when compared to the adjacent tissues (Figure 1A). Consistently, RT-qPCR analysis indicated that SNHG8 expression was up-regulated in all tumor cell lines, including MCF7, Hs-578T, ZR-75-30, HCC1937 cells when compared with normal breast cell MCF-10A. Of note since the expression levels of SNHG8 were higher in MCF7 and ZR-75-30 cells (Figure 1B), we selected these two cell lines for further investigation. The correlation between SNHG8 and miR-634 was analyzed (Figure 1C). All these results indicated that SNHG8 was positively correlated with breast cancer progression.
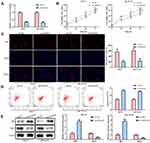
Knockdown of SNHG8 Inhibits Breast Cancer Cells Proliferation, Migration/Invasion, While Accelerating Apoptosis
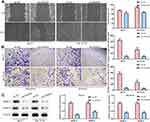
SNHG8 expression in MCF7 and ZR-75-30 cell lines was manipulated using SNHG8 shRNAs in order to explore the function of SNHG8 in breast cancer progression and its oncogenic ability. The knockdown efficiency is validated in Figure 2A. CCK-8 and EdU incorporation assays implied that knockdown of SNHG8 significantly depressed proliferation of MCF7 and ZR-75-30 cell lines (Figure 2B and C). In addition, flow cytometry analysis found that SNHG8 knockdown promoted the apoptosis both in MCF7 and ZR-75-30 cell lines (Figure 2D). At the molecular level, knockdown of SNHG8 significantly decreased the protein expression level of Bax, while increased the Bcl-2 (Figure 2E).
Next, we performed scratch and transwell chamber assays to examine the effect of SNHG8 knockdown on breast tumor cell migration and invasion. As shown in Figure 3A and B, SNHG8 deficiency inhibited cell migration and invasion both in MCF7 and ZR-75-30 cell lines. At the molecular level, the expression levels of migration and invasion-related protein MMP-2 and MMP-9 proteins were significantly down-regulated when SNHG8 was knocked down (Figure 3C). Collectively, these results indicated that SNHG8 functions as a tumor oncogene in breast cancer progression.
miR-634 is a Target Gene of lncRNA SNHG8
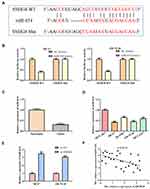
As is known to all, lncRNAs have multiple biological functions through sponging various miRNAs.15 By using bioinformatics analysis (http://starbase.sysu.edu.cn), we notified that a promising binding site existed between lncRNA SNHG8 and miR-634 (Figure 4A). Thus, we investigated whether lncRNA SNHG8 could directly regulate miR-634 expression by using luciferase reporter assays. The results showed that overexpression of miR-634 repressed the luciferase activities containing the wild-type 3ʹ-UTR of SNHG8, but had no obvious effect on mut 3ʹ-UTR of SNHG8 both in MCF7 and ZR-75-30 cell lines (Figure 4B). Additionally, RT-qPCR analysis implied that knockdown of lncRNA SNHG8 significantly up-regulated the miR-634 levels (Figure 4C). In addition, the expression level of miR-634 was decreased in tumor tissues, when compared with normal tissues (Figure 4D). In the meanwhile, similar results were observed in all breast cancer cell lines (Figure 4E). The correlation between miR-634 and ZBTB20 was analyzed (Figure 4F).
Overexpression of miR-634 Suppresses the Progression of Breast Cancer in vitro
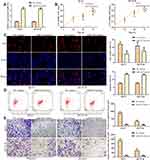
NC mimics and miR-634 mimics were transfected into breast cancer cell lines (MCF7 and ZR-75-30), respectively, so as to investigate the function of miR-634 in breast cancer progression. The transfection efficiency was validated and shown in Figure 5A. As shown in Figure 5B, miR-634 mimics decreased the cell viability. Consistent with these findings, EdU incorporation assay revealed that exogenous miR-634 expression inhibited the proliferation of MCF7 and ZR-75-30 cells (Figure 5C). On the other hand, flow cytometry analysis found that miR-634 mimics promoted the apoptosis of MCF7 and ZR-75-30 cells (Figure 5D). To conclude, miR-634 mimics inhibited cell migration and invasion both in MCF7 and ZR-75-30 cell lines (Figure 5E).
LncRNA SNHG8 Enhances the Progression of Breast Cancer Cell by Regulating miR-634/ZBTB20 Axis
We adopted a miRNA target prediction website (targetscan.org) to screen for miR-634 target genes in order to investigate the detailed mechanism underlying miR-634-regulating breast cancer progression. Among all the targets, we selected ZBTB20, as a potential target of miR-643, because of the important function of ZBTB20 involved in the tumor development and progression. The putative binding sites are shown in Figure 6A. Functionally, miR-643 mimics weakened the activity of ZBTB20 3ʹ-UTR-luciferase reporter. In contrast, mutation in miR-634 binding site in the 3ʹ-UTR of ZBTB20 almost completely offset the repression effect of miR-634 mimics on luciferase activity (Figure 6B), suggesting that this binding site is essential for the function of miR-634. Besides, the expression level of ZBTB20 correspondingly elevated in breast cancer samples, compared with normal tissues (Figure 6C). Meanwhile, similar results were observed in all breast cancer cell lines (Figure 6D). Consistently, RT-qPCR analysis indicated that overexpression of miR-634 significantly inhibited ZBTB20 mRNA and protein expression levels (Figure 6E and F).
To test whether miR-634/ZBTB20 axis was involved in SNHG8-promoted breast cancer progression, miR-634 inhibitors and ZBTB20 shRNAs were transfected into MCF7 cells in the presence with SNHG8 shRNAs. The efficiency of miR-634 inhibition and ZBTB20 knockdown in MCF7 are presented in Figure 7A. CCK-8 and EdU incorporation assays indicated that miR-634 inhibitors induced MCF7 proliferation in the MCF7 cells with lncRNA SNHG8 knockdown, while these induction effects were partially antagonized by knockdown of ZBTB20 (Figure 7B and C). In contrast, knockdown of ZBTB20 recapitulated the miR-634 inhibitors-induced suppression of cell apoptosis in the SNHG8-deficient MCF7 cells (Figure 7D). Coincidence with these results, the cell migration and invasion analyses revealed similar changes as in the proliferation analysis (Figure 7E). Taken together, lncRNA SNHG8 activated breast cancer cell progression by partly targeting the miR-634/ZBTB20 axis.
Discussion
Breast cancer is the most common cancer affecting women and ranks as the second highest cause of cancer-related death among women worldwide.16,17 Over the past few decades, studies have shown that multiple lncRNAs are abnormally expressed in breast cancer.18–20 Therefore, it is important for the identification of breast cancer clinical treatment to dissect the role of lncRNAs in breast cancer progression. In our study, we found that the expression levels of lncRNA SNHG8 were elevated in breast cancer tissues and cell lines. Knockdown of lncRNA SNHG8 decreased the proliferation, migration/invasion, while inducing apoptosis of breast cancer cells.
Broad evidences imply that the lncRNAs play critical role in breast cancer progression. For example, the lncRNA SBF2-AS1 promotes tumorigenesis and progression of breast cancer by regulating the miR-143/RRS1 axis.21 LncRNA LOXL1-AS1 drives breast cancer metastasis by antagonizing miR-708-5p expression and activity.22 On the other hand, lncRNA SNHG8 is found to promote endometrial carcinoma, gastric cancer, liver cancer, as well as the non-small-cell lung cancer,14,23–25 indicating that SNHG8 is essential in the regulation of cancer pathology. In our study, we found that lncRNA SNHG8 was increased in breast cancer samples and cell lines. Our findings broadened the current function of lncRNA SNHG8 in cancer progression.
MiRNAs have become inhibitory and carcinogenic in tumorigenesis.26 Extensive research has shown that a multitude of miRNAs become dysregulated in breast cancer by regulating various pathological changes.27 Therefore, taking in-depth study of these miRNAs provides new opportunities for developing effective techniques to prevent and treat breast cancer. The downstream miRNA-634 in our study was significantly down-regulated in many tumors.28–30It is worth noting that up-regulating the expression level of miR-634 decreases the invasion and proliferation activities of tumor cells effectively.28,30,31 In the present study, we found that miR-634 was a target miRNA of SNHG8. Accordingly, the expression levels of miR-634 were down-regulated and negatively correlated with the SNHG8 in breast cancer tissues and cell lines. Luciferase reporter assays revealed that miR-634 could bind to SNHG8 and decrease its luciferase activity in MCF7 and ZR-75-30 cell lines, thus antagonized the activation effect of SNHG8 on the cancer cell progression. These outcomes indicated that SNHG8 might act as a sponger of miR-634 to accelerate breast cancer cell activation.
On the other hand, miR-634 has been proven to be a new tumor suppressor, which plays an important role in inducing tumor cells apoptosis, especially in liver cancer and hypophysoma.32–34 For example, it up-regulates cell proliferation in nasopharyngeal carcinoma by targeting p-MEK and p-ERK.35 Therefore, screening out the gene target of miR-634 in breast cancer is of great interest. In our study, given that ZBTB20 is a well-known oncogene in other tumors,36 we demonstrated the ZBTB20 was a target gene of miR-634 by bioinformatics and luciferase activity analyses. More importantly, knockdown of ZBTB20 reversed the neutralization effect of miR-634 on the SNHG8 knockdown-induced inhibition of breast cancer cell activation. Therefore, ZBTB20 might function as an output of the SNHG8-miR-634 axis in breast cancer. However, more clinic investigations need to be performed to further reinforce these findings.
In the present study, we demonstrated that the expression level of SNHG8 was upregulated in breast cancer tissues, as well as cell lines. Moreover, knockdown of SNHG8 inhibited breast cancer cell proliferation, migration/invasion while triggered the cell apoptosis. In addition, we found that miRNA-634 was down-regulated in breast cancer tissues, and overexpression of miR-634 partially blocked the SNHG8-induced breast cancer cell progression. Mechanistically, the effect of SNHG8 on breast cancer was partially mediated by miR-634/ZBTB20 axis. Our findings provided a promising mechanism where SNHG8 regulated the progression of breast cancer. SNHG8, as well as miR-634 and ZBTB20, might be adopted as diagnosed biomarkers and therapeutic targets for breast cancer and its associated diseases.
Acknowledgments
This work was supported by the Nature Science Foundation for the Youth of Jiangxi (grant no. 20181BAB205052); The Technology Plan Fund of Jiangxi province department of education project (grant no. GJJ170873); The Open Project of Key Laboratory of Prevention and treatment of cardiovascular and cerebrovascular diseases, Ministry of Education (grants No. XN201809); Chinese medicine project of Jiangxi health and Family Planning Commission (grants No.2014a149).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Konner M. Progress in the treatment of breast cancer. N Engl J Med. 2020;382:e4.
3. Rozeboom B, Dey N, De P. ER+ metastatic breast cancer: past, present, and a prescription for an apoptosis-targeted future. Am J Cancer Res. 2019;9(12):2821–2831.
4. Solinas C, Aiello M, Migliori E, Willard-Gallo K, Emens LA. Breast cancer vaccines: heeding the lessons of the past to guide a path forward. Cancer Treat Rev. 2020;84:101947. doi:10.1016/j.ctrv.2019.101947
5. Banik K, Ranaware AM, Choudhary H, et al. Piceatannol: a natural stilbene for the prevention and treatment of cancer. Pharmacol Res. 2020;153:104635. doi:10.1016/j.phrs.2020.104635
6. Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74.
7. Luo N, Zhang K, Li X, Hu Y. ZEB1 induced-upregulation of long noncoding RNA ZEB1-AS1 facilitates the progression of triple negative breast cancer by binding with ELAVL1 to maintain the stability of ZEB1 mRNA. J Cell Biochem. 2020;121(10):4176–4187. doi:10.1002/jcb.29572
8. Guo S, Jian L, Tao K, Chen C, Yu H, Liu S. Novel breast-specific long non-coding RNA LINC00993 acts as a tumor suppressor in triple-negative breast cancer. Front Oncol. 2019;9:1325. doi:10.3389/fonc.2019.01325
9. Li J, Li Z, Zheng W, et al. LncRNA-ATB: an indispensable cancer-related long noncoding RNA. Cell Prolif. 2017;50.
10. Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–116. doi:10.1093/bib/bbv031
11. Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci (Schol Ed). 2015;7(1):94–108. doi:10.2741/s427
12. Zhang P, Li S, Chen Z, Lu Y, Zhang H. LncRNA SNHG8 promotes proliferation and invasion of gastric cancer cells by targeting the miR-491/PDGFRA axis. Hum Cell. 2020;33(1):123–130. doi:10.1007/s13577-019-00290-0
13. Song Y, Zou L, Li J, Shen ZP, Cai YL, Wu XD. LncRNA SNHG8 promotes the development and chemo-resistance of pancreatic adenocarcinoma. Eur Rev Med Pharmacol Sci. 2018;22(23):8161–8168. doi:10.26355/eurrev_201812_16508
14. Dong J, Teng F, Guo W, Yang J, Ding G, Fu Z. lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR-149-5p and predicts tumor recurrence in hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(5):2262–2274. doi:10.1159/000495871
15. Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem. 2017;118(10):3349–3358. doi:10.1002/jcb.25988
16. Samson M, Porter N, Orekoya O, et al. Progestin and breast cancer risk: a systematic review. Breast Cancer Res Treat. 2016;155(1):3–12. doi:10.1007/s10549-015-3663-1
17. Castaneda SA, Strasser J. updates in the treatment of breast cancer with radiotherapy. Surg Oncol Clin N Am. 2017;26(3):371–382. doi:10.1016/j.soc.2017.01.013
18. Zhang L, Yan X, Yu S, et al. LINC00365-SCGB2A1 axis inhibits the viability of breast cancer through targeting NF-kappaB signaling. Oncol Lett. 2020;19(1):753–762. doi:10.3892/ol.2019.11166
19. Zhu Q, Li Y, Dong X, Yang Y, Wang H, Guo S. Linc-OIP5 loss regulates migration and invasion in MDA-MB-231 breast cancer cells by inhibiting YAP1/JAG1 signaling. Oncol Lett. 2020;19(1):103–112. doi:10.3892/ol.2019.11071
20. Qiu P, Dou Y, Ma LZ, Tang XX, Liu XL, Chen JW. Long non-coding RNA TTN-AS1 promotes the metastasis in breast cancer by epigenetically activating DGCR8. Eur Rev Med Pharmacol Sci. 2019;23(24):10835–10841. doi:10.26355/eurrev_201912_19787
21. Xia W, Liu Y, Cheng T, Xu T, Dong M, Hu X. Down-regulated lncRNA SBF2-AS1 inhibits tumorigenesis and progression of breast cancer by sponging microRNA-143 and repressing RRS1. J Exp Clin Cancer Res. 2020;39(1):18. doi:10.1186/s13046-020-1520-5
22. Dong HT, Liu Q, Zhao T, et al. Long non-coding RNA LOXL1-AS1 drives breast cancer invasion and metastasis by antagonizing mir-708-5p expression and activity. Mol Ther Nucleic Acids. 2020;19:696–705. doi:10.1016/j.omtn.2019.12.016
23. Yang CH, Zhang XY, Zhou LN, et al. LncRNA SNHG8 participates in the development of endometrial carcinoma through regulating c-MET expression by miR-152. Eur Rev Med Pharmacol Sci. 2018;22(6):1629–1637. doi:10.26355/eurrev_201803_14698
24. Liu J, Yang C, Gu Y, et al. Knockdown of the lncRNA SNHG8 inhibits cell growth in Epstein-Barr virus-associated gastric carcinoma. Cell Mol Biol Lett. 2018;23(1):17. doi:10.1186/s11658-018-0070-8
25. Chen C, Zhang Z, Li J, Sun Y. SNHG8 is identified as a key regulator in non-small-cell lung cancer progression sponging to miR-542-3p by targeting CCND1/CDK6. Onco Targets Ther. 2018;11:6081–6090. doi:10.2147/OTT.S170482
26. Jiang C, Chen X, Alattar M, Wei J, Liu H. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015;22(6):291–301. doi:10.1038/cgt.2015.19
27. Mulrane L, McGee SF, Gallagher WM, O’Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73(22):6554–6562. doi:10.1158/0008-5472.CAN-13-1841
28. Fujiwara N, Inoue J, Kawano T, Tanimoto K, Kozaki K, Inazawa J. mir-634 activates the mitochondrial apoptosis pathway and enhances chemotherapy-induced cytotoxicity. Cancer Res. 2015;75(18):3890–3901. doi:10.1158/0008-5472.CAN-15-0257
29. van Jaarsveld MT, van Kuijk PF, Boersma AW, et al. miR-634 restores drug sensitivity in resistant ovarian cancer cells by targeting the Ras-MAPK pathway. Mol Cancer. 2015;14(1):196. doi:10.1186/s12943-015-0464-4
30. Tan Z, Zhao J, Jiang Y. MiR-634 sensitizes glioma cells to temozolomide by targeting CYR61 through Raf-ERK signaling pathway. Cancer Med. 2018;7(3):913–921. doi:10.1002/cam4.1351
31. Peng X, Cao P, He D, et al. MiR-634 sensitizes nasopharyngeal carcinoma cells to paclitaxel and inhibits cell growth both in vitro and in vivo. Int J Clin Exp Pathol. 2014;7(10):6784–6791.
32. Xu D, Yu J, Gao G, Lu G, Zhang Y, Ma P. LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci Rep. 2018;38.
33. Shao B, He L. Hsa_circ_0001742 promotes tongue squamous cell carcinoma progression via modulating miR-634 expression. Biochem Biophys Res Commun. 2019;513(1):135–140. doi:10.1016/j.bbrc.2019.03.122
34. Guo J, Zhang CD, An JX, et al. Expression of miR-634 in gastric carcinoma and its effects on proliferation, migration, and invasion of gastric cancer cells. Cancer Med. 2018;7(3):776–787. doi:10.1002/cam4.1204
35. Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80. doi:10.1016/S0140-6736(19)30956-0
36. Zhao JG, Ren KM, Tang J. Zinc finger protein ZBTB20 promotes cell proliferation in non-small cell lung cancer through repression of FoxO1. FEBS Lett. 2014;588(24):4536–4542. doi:10.1016/j.febslet.2014.10.005
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.