Back to Journals » Cancer Management and Research » Volume 12
LncRNA-SNHG7 Enhances Chemotherapy Resistance and Cell Viability of Breast Cancer Cells by Regulating miR-186
Authors Zhang H, Zhang XY, Kang XN, Jin LJ, Wang ZY
Received 1 July 2020
Accepted for publication 14 September 2020
Published 14 October 2020 Volume 2020:12 Pages 10163—10172
DOI https://doi.org/10.2147/CMAR.S270328
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Hui Zhang,1 Xiao-Yu Zhang,1 Xiao-Ning Kang,2 Li-Jun Jin,1 Zun-Yi Wang1
1Department of Thyroid and Breast III, Cangzhou Central Hospital, Cangzhou, Hebei 061001, People’s Republic of China; 2Department of Ultrasound, Cangzhou Central Hospital, Cangzhou, Hebei 061001, People’s Republic of China
Correspondence: Zun-Yi Wang
Department of Thyroid and Breast III, Cangzhou Central Hospital, Cangzhou, Hebei 061001, People’s Republic of China
Email [email protected]
Background: Clinical tolerance to trastuzumab greatly affects the therapeutic effect in breast cancer (BC). Long-chain non-coding RNA (lncRNA) plays an important role in the development of trastuzumab resistance, in which SNHG7 can promote the epithelial mesenchymal transformation (EMT) of breast cancer cells into, while EMT is related to trastuzumab resistance of breast cancer cells.
Objective: To investigate whether lncRNA-SNHG7 can enhance chemotherapy resistance and cell viability of BC cells by regulating miR-186.
Methods: SK-BR-3 and SNHG7 of HER2+BC cells were induced to enhance the resistance of BC cells to trastuzumab by regulating miR-186, and to regulate the expression levels of SNHG7 and miR-186. The sensitivity of drug-resistant cells to trastuzumab and the changes of cell proliferation, migration, apoptosis, and EMT were measured and verified by tumorigenesis in vivo. The effects of miR-186 on SNHG7 were investigated through rescue experiments; the regulatory relationship between the expression of SNHG7 and miR-186 was verified by the double luciferase reporter (DLR) and the mechanism of SNHG7 was explored.
Results: Down-regulation of SNHG7 or up-regulation of miR-186 could increase the sensitivity of BC cells to trastuzumab, inhibit the proliferation, migration and EMT, and promote apoptosis. Compared with the down-regulation of SNHG7 or miR-186 alone, simultaneous down-regulation of SNHG7 and miR-186 on drug-resistant cells brought notably lower sensitivity to trastuzumab and apoptosis rate, and higher proliferation and apoptosis ability. The DLR showed that miR-186 could specifically inhibit the expression of SNHG7. The results of tumorigenesis in vivo revealed that down-regulation of SNHG7 or up-regulation of miR-186 could improve the therapeutic effect of trastuzumab and reduce the tumor volume, and miR-186 could also antagonize the effect of SNHG7.
Conclusion: Down-regulation of SNHG7-targeted miR-186 can reverse trastuzumab resistance of BC cells, inhibit the proliferation, migration, and EMT levels of BC cells, and promote apoptosis.
Keywords: SNHG7, miR-186, breast cancer, drug resistance
Introduction
Breast cancer (BC) is one of the most common gynecological malignant tumors in the world and the second leading cause of cancer death in women.1,2 In the United States, BC accounted for 30% of all new cancers in 2017.3 Although advances in early diagnosis, surgery, chemotherapy, and other treatment methods have greatly improved the prognosis of BC patients, the survival of them is still not optimistic due to the high heterogeneity of BC cells and the increasing resistance of tumor cells to chemotherapeutic drugs.4 Therefore, the call for understanding the mechanism of BC development and the development of drug resistance, and finding more targets for treating BC is overwhelmingly urgent.
Doxorubicin is one of the drugs of choice for the treatment of BC, however, its clinical application is limited due to its severe side-effects and concomitant drug resistance.5 Of these, trastuzumab is the first approved targeted therapy for HER2+BC, and it is also the first choice for the treatment of this disease. However, with the widespread use of trastuzumab, many patients develop tolerance to trastuzumab, which greatly affects the therapeutic effect of trastuzumab.6,7 Today, increasing evidence has found the role of various molecules in the occurrence of tumor drug resistance with the in-depth understanding of the mechanism of tumor drug resistance.8,9 It is shown that long non-coding RNA (lncRNA) exerts marked effects on the generation of trastuzumab resistance, such as H3K27, and small nucleolar RNA host gene (SNHG) 14.10,11 As another member of the SNHG family, SNHG7 is located on chromosome 9q34.3, which is also associated with the development of tumor resistance. As reported by Chen et al,12 SNHG7 promoted cisplatin resistance in non-small cell lung cancer. Wu et al13 found in their study that SNHG7 was involved in paclitaxel resistance in hypopharyngeal cancer. It still raises questions concerning whether SNHG7 is also implicated in the development of trastuzumab resistance in BC cells, but recent studies have found that SNHG7 can promote epithelial mesenchymal transformation (EMT) of BC cells.14 While, according to Shi et al,15 EMT was associated with trastuzumab resistance in BC cells, suggesting that SNHG7 might also participate in the development of trastuzumab resistance.
In this study, the effect of lncRNA-SNHG7 on chemotherapy resistance and cell viability of BC cells, as well as its mechanism of action were explored through the study of the breast cancer model,16 so as to provide more experimental evidence for clinical search for targets in the treatment of BC.
Materials and Methods
Study Subjects
Two HER2+BC cell lines, SK-BR-3 and AU565, were purchased from American Type Culture Collection (ATCC; Manassas, VA), with Cat. Nos. of ATCC®HTB-30™ and ATCC®CRL-2351™, respectively. All the cells underwent short tandem repeat (STR) authentication, and mycoplasma detection was performed before each cell experiment. The medium of SK-BR-3 cells was McCoy’s 5a (ATCC, 30–2007)+10% fetal bovine serum (ATCC, 30–2020), while that of AU565 cells was RPMI-1640 (ATCC, 30–2001). Both medium contained 1% L-glutamine, 1% sodium pyruvate, 100 U/mL penicillin, and 100 μg/mL streptomycin. The cell culture conditions were 37.0°C, 95% air+5% CO2.
Construction of Drug-Resistant Cell Lines
Two BC cell lines, SK-BR-3 and AU565, were continuously exposed to trastuzumab with increasing concentrations to induce the cells to become resistant to trastuzumab. The initial trastuzumab concentration was 1 μM, and the trastuzumab concentration was increased to 2 μM after 3 months of culture for another 12 months of continuous culture. The drug-resistant cells SK-BR-3/TR and AU565/TR were preserved in a culture medium containing 2 μM trastuzumab. Trastuzumab was purchased from Shanghai Roche Pharmaceutical Co., Ltd. with a specification of 440 mg/branch. Drug resistance index=50% inhibitory concentration (IC50) of drug resistant cells/IC50 of parental cells.
Construction and Transfection of Expression Vectors
SNHG7 low expression vector (si-SNHG7), SNHG7 overexpression vector (sh-SNHG7), miR-186 low expression vector (miR-186-inhibitor), miR-186 overexpression vector (miR-186 mimic), Gal-1-SNHG7 wild type (wt), Gal-1-SNHG7 mutant (mt), and their corresponding control vectors LNC-NC, miR-NC, and Gal-1-NC were designed and synthesized by ThermoFisher Scientific Co., Ltd., Shanghai, China. The cells were digested with trypsin 24 hours before transfection, and transfected with the above expression vectors when the cells reached 80% confluence. Specific procedures were conducted by referring to the kit instructions, and the culture medium was replaced every 6 hours during the 48 hours of culture in a 5% CO2 incubator at 37°C. Quantitative real-time polymerase chain reaction (QRT-PCR) was utilized to detect the transfection results. The cells without any intervention were assigned into the blank group. Lipofectamine TM2000 transfection kit was purchased from Invitrogen Company (Carlsbad, CA), with the Cat. No. 35050.
QRT-PCR
Total RNA was extracted from tissues/cells using TRIzol kit, and the purity, concentration, and integrity were then tested by UV spectrophotometer and agarose gel electrophoresis, at the requirement of Ribosomal RNA 28s:18s≥2 and A260/A280 within 1.8–2.1. The RNA was amplified by one-step method in this study, and the reaction system was as follows: RNA Template: 1 μg, Forward GSP: 0.4 μL, Reverse GSP: 0.4 μL, 2*One-Step Reaction Mix: 10 μL, EasyScript One-Step Enzyme Mix: 0.4 μL, and RNase-free Water was added to complete the reaction volume of 20 μL. The amplification conditions were 40°C for 30 minutes, 94°C for 5 minutes, 94°C for 30 seconds, 60°C for 30 seconds, 72°C for 2 kb/minute, and 72°C for 10 minutes, totaling 42 cycles. GAPDH and U6 were used as the internal reference for lncRNA and miRNA, respectively, and the experiment was run in triplicate. The expression level was calculated by 2−∆∆ct. The TRIzol kit was purchased from Invitrogen Company (US), with the Cat. No. 15596018. The EasyScript One-Step RT-PCR SuperMix kit was acquired from TransGen Biotechnology Co., Ltd. (Beijing, China), with the Cat. No. AE411-02, and the microplate reader was purchased from Flash Spectrum Biotechnology Co., Ltd. (Shanghai, China). Primer sequences are shown in Table 1.
 |
Table 1 Primer Sequences |
CCK8 Cell Proliferation Assay
Cells at logarithmic growth phase were collected, digested with trypsin, and resuspended to 2×104/mL, 100 μL of which was then routinely inoculated into the 96-well plate. At the specific time points, which were 24, 48, 72, and 96 hours, respectively, 200 μL CCK8 mixture (10:1) was added for another 3 hours of culture, and the absorbance value was then measured at the wavelength of 450 nm. The experiment was repeated three times. The CCK8 kit was purchased from Beyotime Biotechnology Co., Ltd., Beijing, China, with the Product No. C0037.
Drug Sensitivity Test
BC cells with a density of 2×103 were routinely inoculated into 96-well plates, and were added with trastuzumab of different concentrations 24 hours later. The sensitivity of the cells to BC was detected by CCK8 method 72 hours later, and the IC50 was calculated. The experiment was performed three times.
Tunel Cell Apoptosis Assay
The apoptosis rate was detected by Tunel method (ThermoFisher Scientific Co., Ltd.). Approximately 5×107 cells/mL were fixed in 4% neutral formaldehyde room temperature for 10 minutes, and 50–100 μL cell suspension was dripped on the slide and dried. After the cells were stained according to the instructions of the kit, the number of apoptotic cells and the total number of cells under 5 high-power microscope were counted under an optical microscope, and the apoptosis rate was calculated. The experiment was run three times.
Cell Migration and Invasion Detection
5×104 cells were seeded into the Transwell upper chamber, and L-15 medium containing 10% FBS was added to the lower chamber. After 24 hours, the cells under the membrane were fixed with 75% methanol, and stained with 0.1% crystal violet. The number of transmembrane cells in five visual fields was counted under 40×10 fields of view, and three parallel experiments were set up. The Transwell chamber and related reagents were obtained from Corning (NY).
EMT Detection
Western Blot Assay
The total protein of cells was extracted by radio-immunoprecipitation assay (RIPA) lysis buffer (Thermo Scientific), and its concentration was detected by bicinchoninic acid (BCA) method (Thermo Scientific). Then, the protein concentration was adjusted to 4 μg/μL, and separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) before transferring to a Polyvinylidene Fluoride (PVDF) membrane. Next, the membrane was dyed with Ponceau S staining solution, soaked in PBST for 5 minutes, and sealed with 5% skim milk powder for 2 hours. Followed by the addition of CD9 and CD63, and MMP-14 (Abcam, Burlingame, CA), primary antibody (1:1000) (Abcam) and an overnight sealing at 4°C. After that, the primary antibody was washed and the horseradish peroxidase-labeled goat anti-rabbit secondary antibody (1:5000) was added, incubated at 37°C for 1 hour, and rinsed 3 times with PBS, 5 minutes each. Finally, the excess liquid on the membrane was blotted with a filter paper, and then an enhanced chemiluminescence (ECL) kit was used to develop in a dark room. The protein bands were scanned and the gray value was analyzed using Quantity One software, wherein the relative expression level of the protein = the gray value of the target protein band/the gray value of the β-Actin protein band. The experiment was run in triplicate. RIPA kit, BCA protein kit, and ECL kit and trypsin were all purchased from Thermo Scientific™ under Cat. Nos. of 89,901, 23,250, 35,055, and 90,058, respectively. Rabbit anti-Vimentin, E-Cadherin monoclonal antibodies, and goat anti-rabbit IgG secondary antibodies were purchased from Abcam, and the Cat. Nos. were ab92547, ab40772, and ab6721, respectively.
DLR Assay
Human embryonic kidney cells 293T were purchased from ATCC (US), with the Cat. No. ATCC®CRL-1573™. After the cells were cultured to logarithmic growth phase, they were transfected with Gal-1-SNHG7 wt, Gal-1-SNHG mt, miR-186 mimic, and miR-NC, respectively. After 48 hours of transfection and the addition of 100 μL pre-mixed Luciferase Assay Reagent II for 2 seconds, the fluorescence intensity was measured by dual luciferase detection system (Beckman CytoFLE S flow cytometer, San Francisco, CA). The experiment was repeated three times.
In vivo Cell Experiment
Forty female thymus free nude BALB/c mice, purchased from Harlan Laboratory (France) with an age of 4–5 weeks and a weight of 20–25 g, were equally and randomly divided into four groups. After 1 week of adaptive feeding with common diet, all mice were subcutaneously injected with 10×106 SK-BR-3/TR cells, which were pre-transfected with si-SNHG7, miR-186 mimic, and si-SNHG7+miR-186 inhibitor (co-transfection), respectively, and the untreated SK-BR-3/TR cells were used as the control. After the tumor grew to 180–200 mm3, 4 mg/kg trastuzumab was injected intraperitoneally once a week for 6 weeks, and normal saline was used as control. One week after the last administration, the mice were killed by cervical dislocation, and the tumor volume was measured. This study was approved by the animal ethics committee of Cangzhou Central Hospital, and carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
Statistical Analysis
The collected data was statistically analyzed using SPSS19.0 software (Asia Analytics Formerly SPSS, China). The measurement data were expressed as mean±standard deviation (mean±SD). The inter-group comparison were performed by the independent sample t-test, while one-way analysis of variance was adopted for multigroup comparisons. LSD test was employed for post hoc test, and Pearson test was used for correlation analysis. P<0.05 denoted a significant difference.
Results
Drug Sensitivity of BC Cells to Trastuzumab
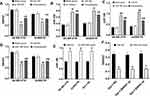
The IC50 values of SK-BR-3 and AU565 against trastuzumab were 1.60 μM and 1.44 μM, while the IC50 values of SK-BR-3/TR and AU565/TR against trastuzumab were 21.44 μM, 20.60 μM, with the resistance index of 13.4 and 14.31, respectively (Figure 1).
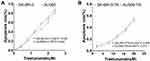 |
Figure 1 Drug sensitivity of BC cells to trastuzumab. (A) Drug sensitivity of parental cells to trastuzumab. (B) Drug sensitivity of drug-resistant cells to trastuzumab. |
Analysis of Transfection results
QRT-PCR results exhibited that si-SNHG7 markedly reduced the expression level of SNHG7 in cells (P<0.05), miR-186 mimic noticeably increased the expression level of miR-186 in cells (P<0.05), and SNHG7 and miR-186 inhibited the expression of each other (Figure 2).
Effects of SNHG7 on Trastuzumab Sensitivity and Cell Viability in Drug-Resistant Cells
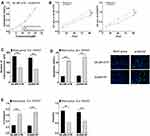
After si-SNHG7 successfully down-regulated the expression level of SNHG7 in drug-resistant cells, the sensitivity of the two drug-resistant cells to trastuzumab increased greatly, and the IC50 values of SK-BR-3/TR and AU565/TR to trastuzumab decreased to 14.74 cm (formerly 21.44 cm) and 13.68 cm (formerly 20.60 cm), respectively. Moreover, we observed that down-regulation of SNHG7 could dramatically reduce the proliferation, migration ability and EMT levels (P<0.05) and promote the apoptosis of drug-resistant cells (P<0.05) (Figure 3).
Effects of miR-186 on Trastuzumab Sensitivity and Cell Viability in Drug-Resistant Cells
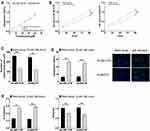
After miR-186 mimic successfully up-regulated the expression of miR-186 in drug-resistant cells, the sensitivity of two drug-resistant cells to trastuzumab increased significantly, and the IC50 of SK-BR-3/TR and AU565/TR to trastuzumab decreased to 12.09 μM (formerly 21.44 μM) and 12.57 μM (formerly 20.60 μM), respectively. What’s more, we noticed that down-regulated miR-186 resulted in dramatically reduced proliferation, migration ability, and EMT levels (P<0.05), as well as enhanced apoptosis of drug-resistant cells (P<0.05) (Figure 4).
MiR-186 Inhibited SNHG7 to Promote Trastuzumab Resistance in BC Cells
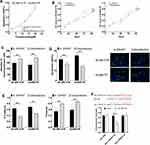
Compared with the si-SNHG7 group, the cells in the co-transfection group were less sensitive to trastuzumab, and the IC50 values of SK-BR-3/TR and AU565/TR for trastuzumab decreased to 17.9 μM (formerly 14.74 μM) and 16.21 μM (formerly 13.68 μM), respectively. In addition, the proliferation, migration, and EMT levels of cells in the co-transfection group elevated (P<0.05), and the apoptosis reduced (P<0.05). The results of the DLR demonstrated that miR-186 was able to target the inhibition of SNHG7 expression (P<0.05) (Figure 5).
Analysis of Tumorigenesis in Mice
The expression level of SNHG7 in tumor tissues in the control group, the co-transfection group, si-SNHG7, and miR-186 mimic group decreased successively (P<0.05), while the expression level of miR-186 increased successively (P<0.05). The expression of SNHG7 and miR-186 showed a significant linear negative correlation (P<0.05), and the tumor volume also decreased in turn (P<0.05) (Figure 6).
Discussion
MiRNAs are a class of short non-coding RNAs approximately 19bp in length that target mRNA through silence-induced complexes with RNA to form a complex and induce its degradation. They usually form a ceRNA network with lncRNA, thus interfering with the function of the protein encoded by the target gene mRNA.17,18 There is evidence showing that lncRNA SNHG14 can target miR-186 to promote cisplatin resistance in colorectal cancer.19 Luo et al20 identified that SNHG7 could target miR-186 to promote the development of BC. In addition, miR-186 has been reported to be involved in EMT21 and inhibit tamoxifen resistance of BC cells.22 We speculate that this may also be one of the mechanisms by which SNHG7 promotes drug resistance in tumor cells. This study, for the first time, found that SNHG7 targeted inhibition of miR-186 promoted BC cell trastuzumab resistance.
In the present study, we constructed two strains of trastuzumab-resistant HER2+BC cells. It was noticed that compared with the parental cells SK-BR-3 and AU565, the IC50 of the resistant cells to trastuzumab increased to 21.44 μM (formerly 1.60 μM), and 20.60 μM (formerly 1.44 μM), respectively, with the drug resistance index of 13.4 and 14.31, presenting moderate drug resistance.23 When SNHG7 was down-regulated in drug-resistant cells, the IC50 of drug-resistant cells to trastuzumab decreased to 14.74 μM and 13.68 μM, respectively, the proliferation and migration capacities of cells decreased obviously, and the apoptosis rate increased. Gao et al24 also reported in their study that SNHG7 can promote the proliferation and invasion of BC cells. Burnett et al25 pointed out that the transformation of HER2+BC induced by EMT to triple-negative breast cancer was an important cause of trastuzumab resistance in HER2+BC. However, in this study, estrogen receptor, progesterone receptor, and proto-oncogene were not detected in the two cell lines, so whether similar changes were to be observed in the cell lines in this study still needs further verification. Nevertheless, the results of the present study show that down-regulation of SNHG7 could inhibit the EMT of drug-resistant cells.
Furthermore, we found that up-regulating the expression of miR-186 in drug-resistant cells could produce similar effects to down-regulating SNHG7, that is, increasing the sensitivity of drug-resistant cells to trastuzumab, inhibiting proliferation, migration, EMT, and promoting apoptosis of BC cells. What’s more, it was observed that miR-186 could antagonize SNHG7, and compared with the down-regulation of SNHG7 alone, the drug-resistant cells that simultaneously down-regulated SNHG7 and miR-186 had significantly lower sensitivity to trastuzumab and higher proliferation and apoptosis ability. Moreover, the DLR indicated that miR-186 could target to inhibit the expression of SNHG7, and hereto we found that SNHG7 targeted to inhibit miR-186 promoted trastuzumab resistance and cell activity in BC cells. In vivo experiments were further carried out for verification. After 6 weeks of treatment with the same dose of trastuzumab, the tumor volume differed significantly by inoculation of BC drug-resistant cells SK-BR-3/TR that had been treated with different treatments. The tumor volume in the transfection group, si-SNHG7, and miR-186 mimic groups decreased in order, and the expression levels of SNHG7 and miR-186 showed a linearly negative correlation. Therefore, in vivo experiments also confirmed that SNHG7 and miR-186 were involved in the resistance of BC cells to trastuzumab, and there was an antagonistic effect between SNHG7 and miR-186. However, there is no report on the effect of SNHG7 regulation of miR-186 on the drug resistance of tumor cells, and this study only used cells and mice as research objects. In addition, the changes of cell cycle progression, cyclin level, and other factors in BC need to be analyzed. Therefore, the results of this study need to be verified by more basic experiments and further clinical trials.
In summary, down-regulation of SNHG7 targeted miR-186 can reverse the resistance of BC cells to trastuzumab, inhibit the proliferation, migration ability, and EMT levels of BC cells, and promote cell apoptosis (Figure 7), which provided an experimental basis for clinical understanding of the mechanism of trastuzumab resistance of BC and the search for new targets for the treatment of BC.
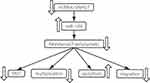 |
Figure 7 SNHG7 targeted miR-186 to reverse trastuzumab resistance of breast cancer cells, inhibit the proliferation, migration, and EMT levels of breast cancer cells, and promote apoptosis. |
Disclosure
The authors report no conflicts of interest for this work.
References
1. R J R, G D R, Koch J, et al. The safety and efficacy of controlled ovarian hyperstimulation for fertility preservation in women with early breast cancer: a systematic review. Human Reproduction. 2017;32(5):1033–1045. doi:10.1093/humrep/dex027
2. H F L, X Z Z, B G L, et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566.
3. Chen Z, Xu L, Shi W, et al. Trends of female and male breast cancer incidence at the global, regional, and national levels, 1990–2017. Breast Cancer Res Treat. 2020;180(2):481–490. doi:10.1007/s10549-020-05561-1
4. Zhang N, Cao C, Zhu Y, et al. Primary breast lymphoma: a single center study. Oncol Lett. 2017;13(2):1014–1018. doi:10.3892/ol.2016.5483
5. Mohammad N, S V S, Malvi P, et al. Strategy to enhance efficacy of doxorubicin in solid tumor cells by methyl-β-cyclodextrin: involvement of p53 and Fas receptor ligand complex. Sci Rep. 2015;5:11853. doi:10.1038/srep11853
6. Palomeras S, Á D-L, Viñas G, et al. Epigenetic silencing of TGFBI confers resistance to trastuzumab in human breast cancer. Breast Cancer Res. 2019;21(1):79. doi:10.1186/s13058-019-1160-x
7. M A S, Gil-Gómez G, Guardia C, et al. Defective cyclin B1 induction in trastuzumab-emtansine (T-DM1) acquired resistance in HER2-positive breast cancer. Clin Cancer Res. 2017;23(22):7006–7019. doi:10.1158/1078-0432.CCR-17-0696
8. Dong H, Hu J, Zou K, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast cancer. Mol Cancer. 2019;18(1):3. doi:10.1186/s12943-018-0931-9
9. Singh S, Chouhan S, Mohammad N, et al. Resistin causes G1 arrest in colon cancer cells through upregulation of SOCS 3. FEBS Lett. 2017;591(10):1371–1382. doi:10.1002/1873-3468.12655
10. Mohammad N, Malvi P, A S M, et al. Cholesterol depletion by methyl-β-cyclodextrin augments tamoxifen induced cell death by enhancing its uptake in melanoma. Mol Cancer. 2014;13(1):204. doi:10.1186/1476-4598-13-204
11. Dong H, Wang W, Chen R, et al. Exosome-mediated transfer of lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int J Oncol. 2018;53(3):1013–1026.
12. Chen K, Abuduwufuer A, Zhang H, et al. SNHG7 mediates cisplatin-resistance in non-small cell lung cancer by activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2019;23(16):6935–6943.
13. Wu P, Tang Y, Fang X, et al. Metformin suppresses hypopharyngeal cancer growth by epigenetically silencing long non-coding RNA SNHG7. Front Pharmacol. 2019;10:143. doi:10.3389/fphar.2019.00143
14. Sun X, Huang T, Liu Z, et al. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the Notch-1 pathway. Eur J Pharmacol. 2019;856:172407. doi:10.1016/j.ejphar.2019.172407
15. Shi J, Li F, Yao X, et al. The HER4-YAP1 axis promotes trastuzumab resistance in HER2-positive gastric cancer by inducing epithelial and mesenchymal transition. Oncogene. 2018;37(22):3022. doi:10.1038/s41388-018-0204-5
16. Muhammad N, Steele R, T S I, et al. Bitter melon extract inhibits breast cancer growth in preclinical model by inducing autophagic cell death. Oncotarget. 2017;8(39):66226. doi:10.18632/oncotarget.19887
17. M D P, Hatzigeorgiou AG. Analyzing Mirna–Lncrna Interactions/Long Non-Coding RNAs. New York, NY: Humana Press; 2016:271–286.
18. Veneziano D, G P M, Di Bella S, et al. Investigating Mirna–Lncrna Interactions: Computational Tools and Resources/Microrna Target Identification. New York, NY: Humana Press; 2019:251–277.
19. Han Y, Zhou S, Wang X, et al. SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis. Biomed Pharmacother. 2020;121:109580. doi:10.1016/j.biopha.2019.109580
20. Luo X, Song Y, Tang L, et al. LncRNA SNHG7 promotes development of breast cancer by regulating microRNA-186. Eur Rev Med Pharmacol Sci. 2018;22(22):7788–7797.
21. Sun W, Zhang Y, Xue P. miR‐186 inhibits proliferation, migration, and epithelial‐mesenchymal transition in breast cancer cells by targeting Twist1. J Cell Biochem. 2019;120(6):10001–10009. doi:10.1002/jcb.28283
22. He M, Jin Q, Chen C, et al. The miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene. 2019;1.
23. Snow K, Judd W. Characterisation of adriamycin-and amsacrine-resistant human leukaemic T cell lines. Br J Cancer. 1991;63(1):17. doi:10.1038/bjc.1991.7
24. Gao Y T, Zhou YC. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur Rev Med Pharmacol Sci. 2019;23(15):6588–6595.
25. Burnett J P, Korkaya H, Ouzounova MD, et al. Trastuzumab resistance induces EMT to transform HER2+ PTEN− to a triple negative breast cancer that requires unique treatment options. Sci Rep. 2015;5:15821. doi:10.1038/srep15821
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.