Back to Journals » Cancer Management and Research » Volume 13
LncRNA SNHG17 Contributes to the Progression of Cervical Cancer by Targeting microRNA-375-3p
Received 26 March 2021
Accepted for publication 3 June 2021
Published 23 June 2021 Volume 2021:13 Pages 4969—4978
DOI https://doi.org/10.2147/CMAR.S312469
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Shuping Cao,1,* Hongxia Li,2,* Lei Li3
1Department of Gynecology, Dongying District People’s Hospital, Dongying, Shandong, People’s Republic of China; 2Department of Obstetrics, Dongying District People’s Hospital, Dongying, Shandong, People’s Republic of China; 3Department of Pathology, Dongying District People’s Hospital, Dongying, Shandong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lei Li
Department of Pathology, Dongying District People’s Hospital, No. 333, Jinan Road, Dongying, Shandong, 257000, People’s Republic of China
Tel/Fax +86-546-8990136
Email [email protected]
Purpose: Cervical cancer is a great threat to women’s health all over the world. Non-coding RNAs performed a wide range of functions. This study aimed to clarify the clinical significance and biological function of lncRNA SNHG17 and miRNA-375-3p (miR-375-3p) in cervical cancer (CC).
Patients and Methods: Blood samples from 124 CC patients and 119 healthy volunteers were collected. The relative expression of SNHG17 and miR-375-3p in CC patient serums and cells was evaluated by quantitative real-time polymerase chain reaction (qRT-PCR). The receiver operating curve (ROC) was plotted for diagnostic value estimation. The CCK-8 and transwell assay were conducted to explore the function of SNHG17 on CC cells. A luciferase reporter assay was carried out to confirm the interaction of SNHG17 and miR-375-3p. Rescue experiments were performed to verify the interaction.
Results: SNHG17 showed an ascending expression while miR-375-3p descended in the serum of CC patients. For SNHG17 and miR-375-3p, respectively, the AUC was 0.863 and 0.869, the sensitivity was 84.7% and 75.8%, and the specificity was 78.2% and 86.6%. Knockdown of SNHG17 inhibited proliferation, migration, and invasion of CC cells. Serum SNHG17 expression was negatively correlated with miR-375-3p expression, and miR-375-3p was the target miRNA of SNHG17. Rescue experiments verified the knockdown of SNHG17 inhibited cell growth through repressing miR-375-3p expression.
Conclusion: SNHG17 and miR-375-3p have the potential to be diagnostic markers for CC. Overexpression of SNHG17 in CC promoted the progression of CC partly via targeting miR-375-3p, implying a novel therapeutic target for CC emerging.
Keywords: lncRNA SNHG17, miR-375-3p, clinical significance, biological function, cervical cancer
Introduction
Cervical cancer (CC) ranked the fourth most common cancer among women internationally, while the second most prevalent malignant neoplasm in low- and middle-income countries.1 In 2018, more than half a million new cases of cervical cancer were diagnosed and 311,365 deaths occurred worldwide due to this malignant disease.2,3 About two-thirds of the patients were diagnosed with locally advanced carcinoma of the cervix, leading to disappointing survival rates in spite of the correct comprehensive treatment.4 Coupled with the metastatic nature of cancer, cervical cancer represents poor outcomes since the moment of diagnosis. Current diagnostic tests for CC are cervical cytology test, human papillomavirus (HPV) primary screening, and contest of these two.5 But cytology test is not sensitive that caused repeat and frequent check just to prevent CC, while the HPV test limited to the virus-infected CC and show a higher proportion of abnormal results than cytology increasing emotional distress.6 Therefore, the development of new biomolecules with specificity and sensitivity for CC is of vital importance to fulfill the unmet need of establishing effective diagnostic approaches in CC.
Non-coding RNAs (ncRNAs) are defined as RNA molecules that do not encode proteins. The extensive functions of ncRNA have become more and more well known, such as the control of chromosome dynamics, RNA editing, inhibition of translation.7 Usually, ncRNAs are categorized as small non-coding RNAs (primarily containing snoRNAs and miRNAs) and long non-coding RNAs (lncRNAs) based on transcript size. lncRNAs are defined as longer than 200 nucleotides non-coding RNAs and compose four-fifths of the total ncRNAs.8 Growing evidence showed lncRNAs and miRNAs possess particular expression in different types of cancer and different pathophysiological stage of tumor growth making them a promising molecular diagnostic tool for a variety of cancers, such as lung cancer,9 breast cancer,10 colorectal cancer,11 thyroid cancer.12 Small nucleolar RNA host gene 17 (SNHG17), one kind of 186-nt lnc RNA with chromosomal location on 20q11.23, has been reported with differential expressions in ovarian cancer,13 oral squamous cell carcinoma,14 lung cancer,15,16 suggesting a potential value as a biomarker in these cancers. Li et al have verified that SNHG17 acted as a miR-485-5p sponge to accelerate lung adenocarcinoma progression.15 In addition, SNHG17 could boost the progression of astrocytoma by binding to miR-876-5p.17 Moreover, SNHG17 was reported to regulate the initiation and progression of colon adenocarcinoma by serves as a sponge for miR-375.18 However, the expression level and potential diagnostic value of SNHG17 in CC have remained unexplored and amorphous.
With the aim to explore the value of SNHG17 and microRNA-375-3p (miR-375-3p), whose deregulation may facilitate pathophysiology of cervical cancer and detect a major subset of high-grade precancerous lesions of cervical scrapes at a good specificity,19 as a diagnostic marker for CC, this study was to determine SNHG17 expression in CC tissues and cell lines and evaluate its diagnostic performance in CC patients. And its biological function in conjunction with miR-375-3p was investigated to help to understand the coherent role of SNHG17 in CC.
Materials and Methods
Serum Collection from Patients and Controls
One hundred and twenty-four CC patients who underwent histopathological examination from March 2018 to December 2020 in Dongying District People’s Hospital were incorporated into the present study. Neither chemotherapy nor radiotherapy was carried out on the patients. And no other treatment history aimed at CC or other infectious diseases was reported by patients. The clinical stages were determined based on the International Federation of Gynecology and Obstetrics criteria (FIGO, 2009). The patient group consisted of 85 cases at stage I–II and 39 cases at III–IV. One hundred and nineteen healthy volunteers with no history of the malignant tumor were enrolled too. The protocol of this study was approved by the institutional ethics review board of Dongying District People’s Hospital (no. 2,018,002). Signed written consent was acquired by the patients before the initiation of any study-related procedure. Blood samples were collected in PAXgene RNA blood collection tubes (BD, USA) from CC patients and healthy participants. All blood samples were separated by centrifugation and sera were collected, aliquoted, and immediately frozen at −80°C for miRNA isolation.
Cell Lines and Transfection
The utilized four cervical cancer cell lines (SiHa, HeLa, C-33A, and MS751) and one normal cervical epithelial cells Ect1/E6E7 were all commercially obtained from Shanghai Kanglang Biological Technology Co., Ltd. All these cells were cultured in RPMI 1640 medium (Gibco by Life Technologies, USA) enriched with 10% (v/v) fetal bovine serum (FBS) and the culture conditions were under a growth temperature 37°C in a 5% CO2 humidified incubator.
Small interfering negative control (si-NC (5ʹ-UUCUCCGAACGUGUCAGGU-3ʹ) and si-SNHG17 (5ʹ-GAUUGUCAGCUGACCUCUGUCCUGU-3ʹ) were purchased from Shanghai Integrated Biotech Solutions Co., Ltd (Shanghai, China). Mimic NC (5ʹ-GCGUGCUUCCGAUUGUUCUGUG-3ʹ), miR-375-3p mimic (5ʹ-UUUGUUCGUUCGGCUCGCGUGA-3ʹ), inhibitor NC (5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ), and miR-375-3p inhibitor (miR inhibitor, 5ʹ-UCACGCGAGCCGAACGAACAAA-3ʹ) were synthesized at GenePharma (Shanghai, China). The transfection was performed according to Lipofectamine 3000 (Invitrogen, USA) transfection manual. Further experiments were performed after transfection for the indicated time.
Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted from serum samples and cells using Trizol reagent (Invitrogen, USA), and reverse-transcribed to complementary DNA (cDNA) using PrimeScript™ RT Master Mix (Takara, Dalia, China) for SNHG17, and PrimeScript Reverse Transcriptase (Takara Bio Inc.) for miR-375-3p. The PCR assay was performed by SYBR Green qPCR Master Mix (Takara, Dalia, China) and a 7500 real-time PCR system (Applied Biosystems, USA). Primers used are supplied by GenePharma (Shanghai, China): SNHG17-fw: 5ʹ-TGCTTGTAAGGCAGGGTCTC-3ʹ; SNHG17-rev: 5ʹ-ACAGCCACTGAAAGCATGTG-3ʹ; GAPDH-fw: 5ʹ- GAAGAGAGAGACCCTCACGCTG-3ʹ; GAPDH-rev:5ʹ-ACTGTGAGGAGGGGAGATTCAGT-3ʹ; miR-375-3p-fw: 5ʹ- GTGCAGGGTCCGAGGT-3ʹ; miR-375-3p-rev: 5ʹ- AGCCGTTTGTTCGTTCGGCT-3ʹ; U6-fw: 5ʹ- CTCGCTTCGGCAGCACA-3ʹ; U6-rev: 5ʹ- AACGCTTCACGAATTTGCGT-3ʹ. The relative expressions of SNHG17 normalized with GAPDH20 and miR-375-3p with U621 were calculated using the 2−ΔΔCt method.
Cell Proliferation Assay
The HeLa and SiHa cell lines were transfected with indicated siRNAs and collected by washing with PBS. Then, cell counting kit-8 (CCK-8; Kumamoto, Japan) was used to access the effect of SNHG17 on CC cell viability. With a density of 3000 cells/well, the cells were suspended in 100 μL fresh medium and seeded into 96-well plates. After 24h incubation at 37°C and 5% CO2 (v/v), 10μL CCK-8 solution was added to the plates followed another 2h incubation at 37°C and 5% CO2and the absorbance was detected at 450 nm (600nm as reference wavelength) according to the microplate reader (Bio-Tek, Winooski, VT, USA) manual. The parallel test was repeated three times.
Cell Migration and Invasion Assay
Cell migration was determined using a 24-well transwell chamber (Corning, New York, USA), and cell invasion using a 24-well transwell chamber (Corning, New York, USA) coated with 50mg/L Matrigel (BD Biosciences). Briefly, indicated-mimics transfected HeLa and SiHa cells in 100 μL serum-free RPMI 1640 medium (Gibco by Life Technologies, USA) were plated in the top chamber, with a final density of 2×105 cell/well. Next, 600 μL complete medium was added to the lower chambers. Twenty-four hours later, the invaded or migrated cells were fixed with methanol, stained with crystal violet.22 The images in five random fields were taken was by an Olympus BX53 microscope carrying DP72 camera (Olympus, Japan) and cell number was counted by Image J software (NIH). Parallel experiments were carried out five times independently.
Apoptosis Assay
Forty-eight-hour post-transfection, SiHa and Hela cells were seeded into 6-well plates at 37°C and 5% CO2 After 12 h incubation, the cells were harvested by trypsinization without EDTA and washed with precooled PBS. Double staining was conducted by Annexin V-FITC/PI cell apoptosis kit (Solarbio, China) and propidium iodide (PI) following the protocol. Then, the cells were analyzed by MoFlo Astrios EQ flow cytometry (Beckman) for the percentage of apoptotic cells. All assay of the samples were in triplicate.
Dual-Luciferase Reporter Assay
The WT- or MUT-SNHG17 containing a miR-375-3p binding sequence was built using QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, USA) and cloned into the pmirGLO Vector (Promega, USA). Then, the construct vectors were transfected into SiHa cells with mimic NC, miR-375-3p mimic, inhibitor NC, or miR-375-3p inhibitor. Subsequently, firefly and renilla (internal reference) luciferase activities were tested with a dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Bioinformatic Analyses of SNHG17 Targeted miR-375-3p
Correlation of SNHG17 with miR-375-3p was analyzed at ENCORI (http://starbase.sysu.edu.cn/).
Statistical Analysis
Statistical analyses were performed in SPSS software (SPSS Inc., Chicago, IL) and GraphPad Prism software (GraphPad Software Inc., San Diego, USA). The data were given as mean ± standard deviation (SD). Two-tailed Student’s t-test or one-way ANOVA with Tukey’s multiple comparisons test was introduced to access the difference among two or three groups. Receiver operating characteristic (ROC) curves were used to evaluate the specificity and sensitivity of SNHG17 for diagnosis of CC. Pearson correlation analysis was used to measure the relationship between SNHG17 and miR-375-3p. P values lower than 0.05 are considered statistically significant.
Results
SNHG17 with Elevated Expression but miR-375-3p with Downward Expression in CC Serum and Cells
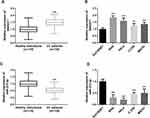
qRT-PCR was performed to determine the relative expression of SNHG17 in CC serum and cell lines. A significantly higher expression of SNHG17 was found in the serum samples obtained from CC patients than those from subjects (p < 0.01, Figure 1A). Similarly, the expressions of SNHG17 in four CC cell lines were significantly higher compared to the normal control cell (all p < 0.001, Figure 1B). Among the four kinds of CC cells, elevated expression of SNHG17 was especially prominent in SiHa and HeLa cells. By contrast, the expression of miR-375-3p in CC samples and cells was significantly decreased when compared with the normal ones (p < 0.001, Figure 1C and D).
Correlation of SNHG17 with Clinical Characteristics of CC Patients
Based on the analysis of the Pearson’s chi-squared test test, the relationship between SNHG17 expression and the clinical-pathological factors of CC patients was explored. The collected samples were divided into a low group (n = 60) and a high group (n = 64) based on the mean value of SNHG17 expression. From the results shown in Table 1, the expression of SNHG17 is affected by FIGO stage (p = 0.023), lymph node metastasis (p = 0.038) and diameter of tumor (p = 0.047). However, no significant correlation was shown between the expression of SNHG17 and age, differentiation degree, or HPV16/18 infection (all p > 0.05).
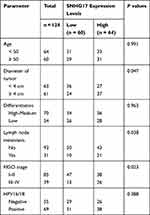 |
Table 1 Correlation Between SNHG17 Expression and Clinicopathological Characteristics in CC Patients |
Diagnostic Value of SNHG17 and miR-375-3p for CC
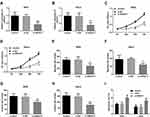
Further, ROC curve analyses were performed to estimate the predictive power of SNHG17 and miR-375-3p in CC diagnosis. As illustrated in Figure 2, both SNHG17 and miR-375-3p are effective in distinguishing CC patients from healthy controls. It was observed that SNHG17 had an AUC of 0.863 (sensitivity = 84.7%, specificity = 78.2%), and miR-375-3p owned an AUC of 0.869 (sensitivity = 75.8%, specificity = 86.6%). These results suggest that SNHG17 and miR-375-3p may be promising diagnostic biomarkers for CC.
Knock-Down of SNHG17 Inhibits CC Cell Proliferation, Migration, and Invasion, and Induces Apoptosis
Since the abnormal expression of SNHG17 in CC serum and cells, a potential biological function of SNHG17 on the progress of CC would have existed. To explore this function, specifically designed siRNAs (si-NC and si- SNHG17) were transfected into SiHa and HeLa cells to downregulate SNHG17 expression (p<0.001, Figure 3A and B). As tested using CCK-8 assays, SNHG17 inhibition obviously repressed the proliferation ability of SiHa and HeLa cells compared to the negative-control treatment (p<0.01, Figure 3C and D). Besides, decreased migrations were detected both in SiHa and HeLa cells through Transwell assay after downregulation of SNHG1723 (p<0.01, Figure 3E and F). There was a similar suppression detected in the invasion experiments (p<0.01, Figure 3G and H). In the apoptosis assay by flow cytometry, the downregulation of SNHG17 dramatically increased the apoptotic cell population (p<0.01, Figure 3I). Based on these results, knockdown of SNHG17 significantly inhibited the proliferative, migratory, and invasive ability of SiHa and HeLa cells, and induced tumor apoptosis cell.
miR-375-3p is a Downstream Target Gene of SNHG17
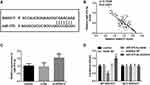
The predicted binding sites from ENCORI are shown in Figure 4A.18 The previous reports supported the association between SNHG17 and miR-375-3p.14,18 Expression of miR-375-3p is negatively correlated with that of SNHG17 in CC serum (r=−0.7049, p<0.0001; Figure 4B). Knockdown of SNHG17 could bring an increase in the expression of miR-375-3p, while si-NC not (p<0.001, Figure 4C). When SiHa cells co-transfected with MUT-SNHG17 and miR-375-3p mimic or miR-375-3p inhibitor, there is no significant change of the relative luciferase activity compared with the cells co-transfected with MUT-SNHG17 and mimic NC and inhibitor NC. However, once the reporter vector carrying the WT-SNHG17 sequence was used, the relative luciferase activity was significantly reduced in the group co-transfected with miR-375-3p mimics and obviously increased in the group co-transfected miR-375-3p inhibitor compared with a miR-NC group (p<0.001; Figure 4D).
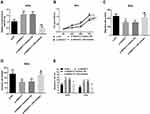
In order to verify the negative correlation between SNHG17 and miR-375-3p, the function of SiHa cells was evaluated after transfected or co-transfected with si-NC, si-SNHG17 and inhibitor NCor miR-375-3p inhibitor. Cells of the si-SNHG17 or si-SNHG17+ miR-375-3p inhibitor exhibited much higher or lower miR-375-3p relative expression than those of respective reference (p<0.001), confirming that SiHa cells were all successfully transfected (Figure 5A). Optical density (OD) values from the CCK-8 assay indicated that inhibition of SNHG17 caused an obvious decrease in cell proliferation, but this decrease can be held back by a miR-375-3p inhibitor (p<0.0001, Figure 5B). Similarly, cell migration and invasion were significantly suppressed when SNHG17 was knockdown, but the suppression could be arrested by a miR-375-3p inhibitor (p<0.001, Figure 5C and D). In addition, miR-375-3p inhibitor can reduce the increase of apoptotic cell population caused by si-SNHG17 (p<0.001, Figure 5E).
Discussion
miRNA as a diagnostic marker for cancers has been studied a lot.24–26 But, in the past, lncRNAs were mostly studied about their nuclear roles. However, with the discovery of some lncRNAs with 3ʹ cleavage and polyadenylation features, lncRNAs present the possibility of being exported to the cytoplasm since resembling the mRNA consensus sequence. These give lncRNAs cytoplasmic functions, for instance, miRNA sponging.27 This large amount of functions, together with a large number of types make lncRNAs represent many abilities, such as acting as oncogenes or tumor suppressors, even diagnostic marker in many diseases. lnc-FAM72D-3 and lnc-EPC1-4 have been identified to be potential candidate biomarkers for HCC diagnosis since it plays a novel role of hepatocarcinogenesis.28 LncRNA lnc-GNAT1-1 was reported to inhibit gastric cancer cell proliferation and invasion in Helicobacter pylori infection.29
In this research, the expression level of SNHG17 and miR-375-3p in CC was studied and their value for CC diagnosis was accessed. SNHG17 turned out to be overexpression but miR-375-3p showed a down-regulated expression in CC patients and cells by qRT-PCR. As to ROC curves, SNHG17 and miR-375-3p represented both good sensitivity and specificity for the diagnosis of CC. With the chi-square test, it is found that SNHG17 overexpression was significantly correlated with FIGO stage, lymph node metastasis, and diameter of tumor in CC patients. So, our results showed SNHG17 and miR-375-3p was a prospective diagnostic marker for CC, implied that SNHG17 and can be developed to diagnostic new molecular markers in CC. Recently, SNHG17 was also reported with overexpression in colorectal cancer tissues and gastric cancer tissues and then can be provided as a prognostic factor for colorectal cancer and gastric cancer.20,30
A mounting number of studies have revealed that aberrant expression levels of ncRNAs may function in cell biological behaviors. To further discover the function of SNHG17 in CC, si-SNHG17 was used to knockdown the expression of SNHG17 in SiHa and HeLa cells, and then proliferation, migration, and invasion of both two kinds of CC cells were inhibited after knockdown of SNHG17. These findings indicate that SNHG17 plays an important role in promoting the development of CC, and may act as biological indicators for the clinical progression and diagnosis of CC. Much recent research in other cancers supported this finding. SNHG17 with high expression in colorectal adenocarcinoma can promote the proliferation and migration of colorectal adenocarcinoma cells.31 Knockdown of SNHG17 decreases prostate cancer cell proliferation, invasion, migration, and epithelial-to-mesenchymal transition (EMT) but strengthened apoptosis, to promote prostate cancer progression.32 Besides, SNHG17 also promotes the progression of breast cancer,33 ovarian cancer,34 and oral squamous cell carcinoma.14 All these studies implied that miR-1298 may play an important role in the progression of various cancers. Since miR-375-3p has been reported to affect the development of HPV16-positive human CC cells and the mechanism has been explored further, this study does not repeat.35
As is known to us, one reason for so many functions of lncRNAs in the cells is that lncRNAs can crosstalk with miRNAs as a competing endogenous RNA (ceRNA).36 Therefore, we searched for the potential targets of SNHG17 in CC and found that miR-375-3p may be a direct target of SNHG17 in CC. This correlation mentioned oral squamous cell carcinoma by Tong’s too.14 In this study, we found miR-375-3p expression was inversely correlated with SNHG17 expression in CC patients. Subsequently, it is validated that miR-375-3p was negatively regulated by SNHG17. Luciferase reporter assay demonstrated that miR-375-3p mimics or inhibitors could obviously influence OD values of WT-SNHG17 plasmid and implied miR-375-3p bind to the 3ʹUTR of SNHG17. Furthermore, rescue experiments suggested that knockdown of SNHG17 inhibited cell growth through repressing miR-375-3p expression at least in part. Succinctly, the results verified that SNHG17- miR-375-3p axis might be the key point of SNHG17 function in CC. On the basis of the researches along with the current study, we reckon that SNHG17 suppresses CC cell proliferation, migration and invasion by targeting miR-375-3p.
The results of this study are promising in a certain extent. However, some limitations also exist: (i) limited to the number of CC cases in our single hospital, further studies getting a larger sample size involved in are recommended; (ii) it is uncertain whether alteration of SNHG17 and miR-375-3p expression in the serum is particular for CC or its certain sub-types, which need additional researches challenging against other cancers or various CC subtypes; (iii) the confirmation of expression level of SNHG17 and miR-375-3p in the CC resected biopsy is needed in the future.
Conclusion
In conclusion, the results in this study indicate that the overexpression of SNHG17 can promote the progression of CC via suppressing the expression of miR-375-3p. SNHG17 and miR-375-3p have obvious clinical significance in CC and can act as candidate indicators of diagnosis and potential therapeutic targets for CC.
Ethics Statement
The protocol of this study was approved by the institutional ethics review board of Dongying District People’s Hospital. Signed written consent was acquired by the patients and healthy volunteers before the initiation of any study-related procedure. Patients and healthy volunteers were informed about the purpose of the study, and that it was conducted in accordance with the Declaration of Helsinki.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Fowler JR, Jack BW. Cervical Cancer. StatPearls. StatPearls Publishing. Copyright © 2020, StatPearls Publishing LLC; 2020.
2. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi:10.1016/S0140-6736(18)32470-X
3. Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8(2):e191–e203. doi:10.1016/S2214-109X(19)30482-6
4. Salunkhe R, Chopra S, Kulkarni S, et al. Outcomes of locally advanced cervical cancer presenting with obstructive uropathy: an institutional audit. Indian J Cancer. 2020;57(4):416–422. doi:10.4103/ijc.IJC_704_18
5. Tsikouras P, Zervoudis S, Manav B, et al. Cervical cancer: screening, diagnosis and staging. J buon. 2016;21(2):320–325.
6. Ngo-Metzger Q, Adsul P. Screening for cervical cancer. Am Fam Physician. 2019;99(4):253–254.
7. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36. doi:10.1038/s41568-020-00306-0
8. Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi:10.3389/fgene.2015.00145
9. Yang B, Zhang L, Cao Y, et al. Overexpression of lncRNA IGFBP4–1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer. 2017;16(1):154. doi:10.1186/s12943-017-0722-8
10. Shima H, Kida K, Adachi S, et al. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res Treat. 2018;170(3):507–516. doi:10.1007/s10549-018-4793-z
11. Li Q-G, Xu X-Q, Zhou D-Y, et al. Long non-coding RNA DILC as a potentially useful biomarker for the diagnosis and prognosis of colorectal cancer. Eur Rev Med Pharmacol Sci. 2019;23(8):3320–3325. doi:10.26355/eurrev_201904_17694
12. Mahmoudian-Sani M-R, Jalali A, Jamshidi M, et al. Long non-coding RNAs in thyroid cancer: implications for pathogenesis, diagnosis, and therapy. Oncol Res Treat. 2019;42(3):136–142. doi:10.1159/000495151
13. Pan X, Guo Z, Chen Y, et al. STAT3-induced lncRNA SNHG17 exerts oncogenic effects on ovarian cancer through regulating CDK6. Mol Ther Nucleic Acids. 2020;22:38–49. doi:10.1016/j.omtn.2020.08.006
14. Tong F, Guo J, Miao Z, Li Z. LncRNA SNHG17 promotes the progression of oral squamous cell carcinoma by modulating miR-375/PAX6 axis. Cancer Biomark. 2021;30(1):1–12. doi:10.3233/CBM-191070
15. Li W, Zheng Y, Mao B, Wang F, Zhong Y, Cheng D. SNHG17 upregulates WLS expression to accelerate lung adenocarcinoma progression by sponging miR-485–5p. Biochem Biophys Res Commun. 2020;533(4):1435–1441. doi:10.1016/j.bbrc.2020.09.130
16. Xu T, Yan S, Jiang L, et al. Gene amplification-driven long noncoding RNA SNHG17 regulates cell proliferation and migration in human non-small-cell lung cancer. Mol Ther Nucleic Acids. 2019;17:405–413. doi:10.1016/j.omtn.2019.06.008
17. Du F, Hou Q. SNHG17 drives malignant behaviors in astrocytoma by targeting miR-876-5p/ERLIN2 axis. BMC Cancer. 2020;20(1):839. doi:10.1186/s12885-020-07280-8
18. Liu J, Zhan Y, Wang J, Wang J, Guo J, Kong D. lncRNA-SNHG17 promotes colon adenocarcinoma progression and serves as a sponge for miR-375 to regulate CBX3 expression. Am J Transl Res. 2020;12(9):5283–5295.
19. Babion I, Snoek BC, Novianti PW, et al. Triage of high-risk HPV-positive women in population-based screening by miRNA expression analysis in cervical scrapes; a feasibility study. Clin Epigenetics. 2018;10(1):76. doi:10.1186/s13148-018-0509-9
20. Ma Z, Gu S, Song M, et al. Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer. Mol Biosyst. 2017;13(11):2350–2361. doi:10.1039/C7MB00280G
21. Wu Y, Sun X, Song B, Qiu X, Zhao J. MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med. 2017;6(7):1686–1697. doi:10.1002/cam4.1110
22. Jules J, Maiguel D, Hudson BI, Oury TD. Alternative splicing of the RAGE cytoplasmic domain regulates cell signaling and function. PLoS One. 2013;8(11):e78267. doi:10.1371/journal.pone.0078267
23. van de Merbel AF, van der Horst G, Buijs JT, van der Pluijm G. Protocols for migration and invasion studies in prostate cancer. Methods Mol Biol. 1786;67–79.
24. McGuire A, Brown JAL, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34(1):145–155. doi:10.1007/s10555-015-9551-7
25. Solé C, Moliné T, Vidal M, Ordi-Ros J, Cortés-Hernández J. An exosomal urinary miRNA signature for early diagnosis of renal fibrosis in lupus nephritis. Cells. 2019;8(8). doi:10.3390/cells8080773
26. Tutar Y. Editorial (Thematic issue: “miRNA and cancer; computational and experimental approaches”). Curr Pharm Biotechnol. 2014;15(5):429. doi:10.2174/138920101505140828161335
27. Mishra PK, Nemer G. Editorial: the non-coding genome and cardiovascular disease. Front Cardiovasc Med. 2019;6:98. doi:10.3389/fcvm.2019.00098
28. Yao Z, Jia C, Tai Y, et al. Serum exosomal long noncoding RNAs lnc-FAM72D-3 and lnc-EPC1-4 as diagnostic biomarkers for hepatocellular carcinoma. Aging. 2020;12(12):11843–11863. doi:10.18632/aging.103355
29. Liu L, Shuai T, Li B, Zhu L, Li X. Long non-coding RNA lnc-GNAT1-1 inhibits gastric cancer cell proliferation and invasion through the Wnt/β-catenin pathway in Helicobacter pylori infection. Mol Med Rep. 2018;18(4):4009–4015. doi:10.3892/mmr.2018.9405
30. Chen -L-L, He J, Qiu X-T, Yu J, Wang Z-M. The prognostic roles of long non-coding RNA SNHG17 in the patients with gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23(3):1063–1068. doi:10.26355/eurrev_201902_16994
31. Liu Y, Li Q, Tang D, et al. SNHG17 promotes the proliferation and migration of colorectal adenocarcinoma cells by modulating CXCL12-mediated angiogenesis. Cancer Cell Int. 2020;20(1):566. doi:10.1186/s12935-020-01621-0
32. Wu G, Hao C, Qi X, et al. LncRNA SNHG17 aggravated prostate cancer progression through regulating its homolog SNORA71B via a positive feedback loop. Cell Death Dis. 2020;11(5):393. doi:10.1038/s41419-020-2569-y
33. Du Y, Wei N, Hong J, Pan W. Long non-coding RNASNHG17 promotes the progression of breast cancer by sponging miR-124-3p. Cancer Cell Int. 2020;20(1):40. doi:10.1186/s12935-020-1129-y
34. Zheng Z-J, Liu Y, Wang H-J, Pang -W-W, Wang Y. LncRNA SNHG17 promotes proliferation and invasion of ovarian cancer cells through up-regulating FOXA1. Eur Rev Med Pharmacol Sci. 2020;24(18):9282–9289. doi:10.26355/eurrev_202009_23010
35. Yu X, Zhao W, Yang X, Wang Z, Hao M. miR-375 affects the proliferation, invasion, and apoptosis of HPV16-positive human cervical cancer cells by targeting IGF-1R. Int J Gynecol Cancer. 2016;26(5):851–858. doi:10.1097/IGC.0000000000000711
36. Kim J, Abdelmohsen K, Yang X, et al. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016;44(5):2378–2392. doi:10.1093/nar/gkw017
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.