Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA SNHG16 Regulates the Progress of Acute Myeloid Leukemia Through miR183-5p–FOXO1 Axis
Authors Yang R, Ma D, Wu Y, Zhang Y, Zhang L
Received 17 April 2020
Accepted for publication 25 August 2020
Published 17 December 2020 Volume 2020:13 Pages 12943—12954
DOI https://doi.org/10.2147/OTT.S258684
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
This paper has been retracted.
Ru Yang, 1 Dong Ma, 2 Yanwei Wu, 3 Yingzi Zhang, 4 Lina Zhang 5
1Henan Key Laboratory of Neurorestoratology, First Affiliated Hospital of Xinxiang Medical University, Weihui, Henan Province, People’s Republic of China; 2Hematology Laboratory, First Affiliated Hospital of Xinxiang Medical University, Weihui, Henan Province, People’s Republic of China; 3Clinical Laboratory, First Affiliated Hospital of Xinxiang Medical University, Weihui, Henan Province, People’s Republic of China; 4Department of Blood Transfusion, First Affiliated Hospital of Xinxiang Medical University, Weihui, Henan Province, People’s Republic of China; 5Central Laboratory, First Affiliated Hospital of Xinxiang Medical University, Weihui, Henan Province, People’s Republic of China
Correspondence: Ru Yang
Henan Key Laboratory of Neurorestoratology, First Affiliated Hospital of Xinxiang Medical University, Weihui 453100, Henan Province, People’s Republic of China
Email [email protected]
Purpose: At present, there is a lack of precise knowledge on acute myeloid leukemia (AML) at the molecular level, and understanding its occurrence at the genetic level is conducive to the development of targeted therapies. Therefore, in this study the relationship between the lncRNA SNHG1 –miR183-5p–FOXO1 axis and AML was explored.
Methods: Expression of lncRNA SNHG16 and miR183-5p was quantified by quantitative real-time PCR, and the level of FOXO1 and other proteins was measured by Western blot. Expression vectors of lncRNA SNHG16, miR183-5p, and FOXO1 were constructed to assess effects of the three on cell proliferation and apoptosis. MTT reduction assays were employed for cell proliferation, flow cytometry for cell cycle and apoptosis, and dual luciferase–reporter assays for the targeting relationship between lncRNA SNHG16 and miR183-5p and miR183-5p and FOXO1.
Results: lncRNA SNHG16 was highly expressed in peripheral blood/leukemia cell lines of patients with AML compared with normal human peripheral blood/peripheral blood mononuclear cells. miR183-5p was the target of lncRNA SNHG16 and FOXO1 the target gene of miR183-5p rather than lncRNA SNHG16. Absence of lncRNA SNHG16 led to upregulation of miR183-5p, promotion of apoptosis, and inhibition of proliferation. Suppression of miR183-5p accelerated cell proliferation and hindered apoptosis. miR183-5p negatively regulated FOXO1, and FOXO1 promoted proliferation and inhibited apoptosis. Inhibition of miR183-5p counteracted the changes caused by lncRNA SNHG16 absence.
Conclusion: lncRNA SNHG16 regulates the progress of AML via the miR183-5p–FOXO1 axis.
Keywords: acute myeloid leukemia, miR183-5p, lncRNA SNHG16, FOXO1
Introduction
Malignant proliferation of progenitor cells and poor expansion of immature myeloblasts caused by gene mutations are the underlying causes of acute myeloid leukemia (AML).1 AML prognosis can be affected by various factors, and the 5-year survival is 27.4%.2 Expectant mothers of advanced reproductive age are more likely to give birth to babies with AML,3 and maternal intake of coffee and tea is also a risk factor for childhood AML.4 In addition, dieldrin, formaldehyde, age, sex, and race are all related to this disease.1,5,6 The molecular pathogenesis of AML remains unknown, and understanding its development from the genetic level contributes to the progress of targeted therapies.
lncRNA is an important component of epigenetic regulation. There are sequence fragments that can bind to downstream target genes in the 3ʹUTR of lncRNA, through which the expression of those target genes is regulated. lncRNA SNHG16 is closely related to tumorigenesis and apparently regulates downstream genes by pairing with various nucleic acid sequences. In pancreatic cancer, lncRNA SNHG16 binds directly to miR128 to upregulate cancer-cell viability and HMGB1 expression.7 It can also promote malignant behavior by gastric cancer cells by regulating the miR135a–JAK2–STAT3 axis.8 The regulatory axis composed of lncRNA SNHG16, miR4500, and STAT3 accelerates the growth of liver cancer cells and the progression of liver cancer.9 The active SNHG16–miR146a axis plays a role in stimulating the metastasis and growth of non–small cell lung cancer carcinoma, leading to poor prognosis.10 In addition, lncRNA SNHG16 has been reported to be involved in the pathogenesis of acute lymphoblastic leukemia.11 These studies all illustrate that lncRNA SNHG16 can regulate the development and progression of tumors and other diseases by binding to miRNA. miR183-5p, a mature spliceosome of miR183 abnormally expressed in various diseases,12–14 acts as both a tumor promoter to induce tumor formation and a tumor suppressor to prevent tumor progression.15–18 FOXO1 contains four functional domains — nuclear localization signal, transactivation (TA), nuclear export signal, and Forkhead (FKH) — and participates in cell physiological and biochemical processes of apoptosis, metabolism, and antioxidant stress.19 Therefore, uncontrolled expression of FOXO1 may induce an increase in malignant cell behavior.
In this study, we found that lncRNA SNHG16 was upregulated in AML by comparing peripheral blood samples of healthy participants and AML patients. After downregulating lncRNA SNHG16 of a leukemia cell line by siRNA, miR183-5p was upregulated and FOXO1 downregulated. In addition, downregulating miR183-5p induced upregulation of FOXO1. These findings suggest that there seems to be an association between lncRNA SNHG16, miR183-5p, and FOXO1, though whether they are related to AML is unknown. Therefore, in this study we explored mechanisms of lncRNA SNHG16, miR183-5p, and FOXO1 in AML.
Methods
Sample Collection
Peripheral blood samples were collected from 76 patients with AML diagnosed at the First Affiliated Hospital of Xinxiang Medical University (42 males and 34 females). Another 68 healthy individuals undergoing checkups in the same period were enrolled as a control group: 39 males and 29 females. The inclusion criterion was AML diagnosis. Exclusion criteria were mental diseases or other tumors, previous surgery, chemotherapy, radiotherapy, or antibiotic therapy. Patients were fully informed, and the study was approved by the ethics committee of the hospital. Peripheral blood was centrifuged and the supernatant collected and stored at −80°C. All participants signed an informed-consent form and the study was conducted in accordance with the Declaration of Helsinki.
Cell Culture and Transfection
Leukemia cell lines (THP1, HL60, Kasumi 3, and AML139) and human peripheral blood mononuclear cells (PBMCs) were purchased from ATCC. Cells were cultured in a 37°C/5% CO2 incubator containing 1640 medium (Hyclone) + 10% FBSFBS solution (Gibco) + 1% penicillin–streptomycin solution (100X, Solarbio). Subsequent experiments were carried out after cell coverage had reached 80%–90%. The day before transfection, the medium was replaced with FBS-free medium. On the day of transfection, cells (105 cells/well) were inoculated into a six-well plate. SNHG16 siRNA, FOXO1 siRNA, FOXO1 pcDNA, miR183-5p inhibitor, miR183-5p mimics, NC siRNA, NC inhibitor, and NC mimic vectors were purchased from Shanghai Sangon Bioengineering. Cell lines were transfected using Lipofectamine 2000 (Invitrogen, USA) according to the instructions. After transfection for 8 hours, cells were cultured in fresh medium at 37°C/5% CO2.
Total RNA Extraction and Quantification by qPCR
Total RNAs collected from sera of 76 patients with AML and 68 healthy individuals were extracted with an miRNeasy serum/plasma kit (217184; Qiagen, Germany), and those from leukemia cell lines and human PBMCs were extracted by Trizol. Optical density (OD) of total RNAs at 260–280 nm was read with ultraviolet spectrophotometry, and those with OD260/OD280>1.8 were taken for subsequent qPCR detection. Reverse transcription and PCR amplification were carried out with a FastKing one-step reverse-transcription fluorescence quantitative kit (FP314; Tiangen, Beijing) and ABI Prism 7000 (Applied Biosystems, USA), respectively. lncRNA SNHG16, miR183-5p, and FOXO1 mRNA primers were designed and synthesized at Sangon Bioengineering. qPCR reactions were performed with reference to the kit manual (total reaction volume 50 μL): 1.25 μL upstream primer, 1.25 μL downstream primer, 1 μL probe, RNA template 10 pg/g, 5 μL of 50× Rox reference dye, and finally add RNase-Free ddH2O to 50 μL. The reaction process comprised one cycle of reverse transcription at 50°C for 30 minutes, one cycle of predenaturation at 95°C for 3 minutes, 40 cycles of denaturation at 95°C for 15 seconds and annealing at 60°C for 30 seconds. Results obtained were analyzed with the ABI Prism 7000. U6 and GAPDH served as internal reference genes, and data were normalized by 2–ΔΔCt (Table 1).
 |
Table 1 Primer sequences |
Protein Expression on Western Blot
Protein expression was quantified by Western blot 96 hours after transfection. A 1mL protein extract (cell lysate:protease inhibitors:phosphatase inhibitor 98:1:1, v:v:v) was repeatedly pipetted until complete lysis of cells. Cells were centrifuged at 1.2×104 rpm for 15 minutes to obtain supernatant. Afterward, protein was separated with SDS-PAGE, transferred to a nitrocellulose (NC) membrane, and placed at room temperature for 1 hour (blocked by 5% skim milk–PBSsolution). Subsequently, FOXO1, caspase 3, caspase 9, Bax, Bcl2, and β-actin primary antibodies were added and left to stand at 4°C overnight. The NC membrane was washed with PBS three times, then goat antirabbit secondary antibody (HRP conjugate) was added and the mixture allowed to stand for 1 hour at room temperature. Finally, the membrane was washed with PBS and visualized with enhanced chemiluminescence. β-actin was taken as the internal reference, and the relative expression level of the test protein = gray value of the test band/gray value of the β-actin band. FOXO1, caspase 3, caspase 9, Bax, Bcl2, β-actin primary antibodies, and goat antirabbit secondary antibody (HRP conjugate) were all purchased from Abcam.
Cell Apoptosis and Cycle on Flow Cytometry
The cell suspension was prepared with trypsin and the number of cells controlled to 106. Cells were immobilized in 70% ice-cold ethanol for 30 minutes, keeping the ambient temperature at 4°C. The ethanol solution was then removed and the cells incubated in annexin V–FITC–7AAD solution. A FACScan flow cytometer (Becton Dickinson) was employed to determine the apoptosis. Cell-cycle detection was operated similarly to apoptosis, only the annexin V–FITC–7AAD solution was replaced with propidium iodide (50 ng/mL)–RNase (0.2 mg/mL) solution and cells treated at room temperature for 30 minutes. Forward and side scattering were measured. A pulse width–area graph was used to construct a gate and an individual cell group selected. The gate was then applied to the scattering pictures, and cell fragments were excluded. Two gates were combined and applied to the PI histogram. Percentage indicates the proportion of cells in each phase. The apoptotic ratio is the proportion of of cells in Q2 and Q3 quadrants to the total number of cells in the gated graph.
Cell Viability on MTT Reduction Assay
Transfected cells were trypsinized. After centrifugation to remove trypsin, fresh medium was added and pipetted to prepare the cell suspension. Cells were inoculated into four 96-well plates at 5×103/100 μL/well, with three wells per group. One plate was taken out every 24 hours. Plates had 5 mg/mL MTT solution (10 μL/well) added and were cultured for 1 hour. Afterward, the medium was removed and OD570 was read with a microplate reader. The experiment was repeated three times to construct the cell viability–time curve.
Targeting Relationships Between lncRNA SNHG16 and miR183-5p and miR183-5p and FOXO1 by Dual Luciferase–Reporter Assay
After prediction of binding sites between miR183-5p and FOXO1 or SNHG16 with TargetScan 7.2 and StarBase 2.0, site mutations were performed and expressed as FOXO1-mut and SNHG16-mut respectively. FOXO1-mut and SNHG16-mut indicated that no sites bound to miR183-5p, and FOXO1-wt and SNHG16-wt indicated sites bound to miR183-5p. PmirGLO-SNHG16-wt, pmirGLO-FOXO1-wt, pmirGLO-SNHG16-mut, and pmirGLO- FOXO1-mut vectors were constructed and cotransfected into cells with miR195-5p mimics and NC mimics. Luciferase activity was detected 48 hours after transfection with a dual luciferase reporter–gene kit (Promega). In addition, FOXO1pcDNA (Tiangen) was used to upregulate FOXO1 in leukemia cell lines. FOXO1pcDNA cell lines cotransfected with PmirGLO-SNHG16-wt or pmirGLO-SNHG16-mut and dual luciferase–reporter assay was employed to determine fluorescence intensity in each group.
Statistical Analysis
Data collected were entered into SPSS20.0 and GraphPad Prism 6.0 for statistical analysis. Each experiment was repeated three times.Measurement data are expressed as mean ± SD. Comparison between the two groups was conducted with independent-sample t-tests, comparison among multiple groups with one-way ANOVA, and post hoc pairwise comparisons with Fisher’s least significant difference t-tests. Pearson analysis was adopted for correlations between lncRNA SNHG16 and miR183-5p and miR183-5p and FOXO1. All data were analyzed with two-tailed tests, with 95% was taken as the CI. Statistical significance was P<0.05.
Results
High Expression of lncRNA SNHG16 in AML
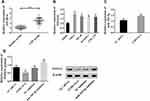
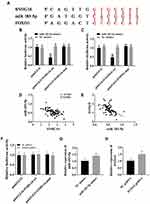
Peripheral blood samples from 76 patients with AML and 68 healthy people were collected. qPCR was employed to quantify lncRNA SNHG16 in peripheral blood and leukemia cell lines. Serum lncRNA SNHG16 in patients with AML was higher than healthy individuals, and that in leukemia cell lines was also higher than that in normal PBMCs (Figure 1A and B). THP1 cells had the highest lncRNA SNHG16 level, so they were selected as the research object. After transfection with SNHG16 siRNA, miR183-5p in THP1 cells was upregulated (Figure 1C), while FOXO1 was downregulated (Figure 1D). After transfection with miR183-5p inhibitor, FOXO1 was upregulated in THP1 cells (Figure 1D). These results indicated that lncRNA SNHG16 was highly expressed and might regulate miR183-5p and FOXO1 expression in AML.
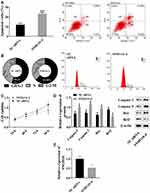
lncRNA SNHG16 Promoted Cell Proliferation and Inhibited Apoptosis
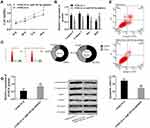
In this study, siRNA interfered with the expression of lncRNA SNHG16 in THP1 cells to explore the function of lncRNA SNHG16 in AML. Flow cytometry was adopted to detect apoptosis and cell cycle, MTT to determine cell viability, and Western blot to quantify expression levels of caspase 3, caspase 9, Bax, and Bcl2 proteins (Figure 2). Absence of lncRNA SNHG16 led to increased apoptosis ratio (Figure 2A), upregulation of caspase 3, caspase 9, and Bax and downregulation of Bcl2 (Figure 2D). Absence of lncRNA SNHG16 resulted in a decrease in S-phase cells and an increase in G0/G1-phase cells (Figure 2B), and cell viability was significantly lowered (Figure 2C). In addition, it upregulated miR183-5p and downregulated FOXO1 (Figure 1). Therefore, lncRNA SNHG16 might accelerate cell proliferation and inhibit apoptosis by regulating miR183-5p and FOXO1.
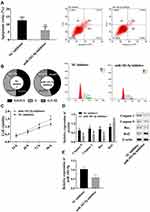
miR183-5p Promoted Apoptosis
Due to the absence of lncRNA SNHG16 increasing miR183-5p, we inhibited miR183-5p in AML cells to observing the effects. Flow cytometry, MTT, and Western blot were used for this part (Figure 3). Compared with the NC inhibitor group, suppression of miR183-5p reduced apoptosis ratio (Figure 3A), and downregulated caspase 3, caspase 9, and Bax and upregulated Bcl2 (Figure 2D). Suppression of miR183-5p promoted S-phase cells, but inhibited G0/G1-phase cells (Figure 3B), and cell viability was significantly lowered (Figure 3C). Suppression of miR183-5p also upregulated FOXO1 (Figure 1D). Overall, miR183-5p inhibited cell proliferation, but promoted apoptosis. miR183-5p was not only regulated by lncRNA SNHG16 but also participated in the regulation of FOXO1.
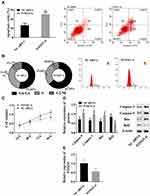
lncRNA SNHG16–miR183-5p Axis Regulated Cell Processes via FOXO1
In view of the changes in FOXO1, we investigated the association between FOXO1 and AML. Compared with the NC siRNA group, absence of FOXO1 led to increased apoptosis ratio (Figure 4A), upregulation of caspase 3, caspase 9, and Bax, and downregulation of Bcl2 (Figure 4D). Absence of FOXO1 also resulted in reduced S-phase cells and increased G0/G1-phase cells (Figure 4B), and cell viability was significantly lowered (Figure 4C). Therefore, FOXO1 promoted cell proliferation and inhibited apoptosis. The lncRNA SNHG16–miR183-5p axis regulated cell processes via FOXO1.
Targeted Relationships among lncRNA SNHG16, miR183-5p, and FOXO1
StarBase demonstrated that there were binding sites between lncRNA SNHG16 and miR183-5p, while TargetScan predicted the existence of pairing sites between miR183-5p and FOXO1 (Figure 5A). Pearson analysis showed that lncRNA SNHG16 was negatively correlated with miR183-5p and miR183-5p negatively correlated with FOXO1 (Figure 5C and E). Dual luciferase–reporter gene assays confirmed that there were targeted relations between lncRNA SNHG16 and miR183-5p (Figure 5B) and miR183-5p and FOXO1 (Figure 5D), indicating that lncRNA SNHG16 targeted miR183-5p, while miR183-5p targeted FOXO1. Quantification of lncRNA SNHG16 and FOXO1 in leukemia cell lines is shown in Figure 5G and H, respectively. Figure 5F indicates that lncRNA SNHG16 failed to bind to FOXO1.
Downregulation of miR183-5p Rescued lncRNA SNHG16-Induced Changes in Leukemia Cell Lines
SNHG16 siRNA and SNHG16 siRNA + miR183-5p inhibitor were used to transfect THP1 cells and corresponding cell changes observed. In SNHG16 siRNA + miR183-5p inhibitor group, cell viability and S-phase cells increased, but cell apoptosis and G0/G1-phase cells decreased. Meanwhile, caspase 3, caspase 9, and Bax were downregulated, and FOXO1 and Bcl2 were upregulated. Those results were comparable with the SNHG16 siRNA group. Therefore, suppression of miR183-5p offset FOXO1 downregulation caused by SNHG16 siRNA, promoted cell proliferation, and inhibited apoptosis (Figure 6).
Discussion
A possible pathogenesis of AML is abnormal apoptosis and proliferation of normal cells in the bone marrow triggered by phenotypic changes in genes or environment hindering the production of normal blood cells. As a common acute leukemia in adults, AML needs to be treated precisely. Epigenetic regulation can be directly involved in gene regulation without changing its sequence, which makes the related ncRNAs become a hot spot in the study of the pathogenesis of various diseases.
Expression of lncRNA SNHG16 in cancer/adjacent tissues was determined, and it turned out that lncRNA SNHG16 was highly expressed in AML. In addition, after suppressing its expression, miR183-5p expression was upregulated and FOXO1 protein expression downregulated. StarBase predicted that lncRNA SNHG16 had matching sequences with miR183-5p, and TargetScan predicted that miR183-5p had binding sites with FOXO1. Therefore, we speculated that abnormal levels of lncRNA SNHG16, miR183-5p, and FOXO1 might be related to the development of AML.
Inhibitory expression vectors of lncRNA SNHG16 and miR183-5p were constructed to explore their functions in AML. Changes in leukemia cells were observed by flow cytometry and MTT, and the expression of related sequences and proteins detected by qPCR and Western blot. The results showed that absence of SNHG16 inhibited cell viability and FOXO1 expression. Most cells were arrested in the G1 phase, accompanied by cell apoptosis andincreased miR183-5p. At the same time, suppressing miR183-5p elevated cell viability and FOXO1 expression, inhibited cell apoptosis, and significantly increased S-phase cells. These results indicated that SNHG16 might affect cell apoptosis and proliferation through miR183-5p–mediated FOXO1.
In order to prove this, FOXO1 expression was suppressed with siRNA. Liu et al revealed that inhibition of FOXO1 expression induced apoptosis in leukemia cells,20 and we found that absence of FOXO1 arrested the cell cycle in the G1 phase, promoted the expression of caspase 3, caspase 9, and Bax proteins, and eventually led to increased apoptosis and decreased cell viability. These results suggested that lncRNA SNHG16 regulated cell proliferation and apoptosis through miR183-5p–mediated FOXO1. FOXO1, an important cell-growth regulator,21 is closely related to cell death and extracellular matrix remodeling.22 As mentioned, FOXO1 is a protein involved in cell apoptosis. When being upregulated by lncRNA SNHG16–miR183-5p, FOXO1 regulates transcription and expression of downstream target proteins, such as Bcl2, Bad, Bim, and TRAIL,23,24 which affects the apoptosis process directly. In addition, FOXO1 can interact with β-catenin to activate the Notch pathway, resulting in malignant proliferation of hematopoietic stem cells.25 In summary, the lncRNA SNHG16–miR183-5p/FOXO1 axis regulated the apoptosis and cell cycle of AML cells.
This study discussed regulation mechanisms of the lncRNA SNHG16–miR183-5p–FOXO1 axis involved in AML, and found that lncRNA SNHG16 promoted cell proliferation and inhibited apoptosis by regulating miR183-5p and FOXO1, which resulted in malignant progression of AML. Although we described the mechanism of the lncRNA SNHG16–miR183-5p–FOXO1 axis from the perspective of molecular biology, its clinical value needs to be further studied. In future experiments, we will explore its influence on drug resistance of AML cells and its relationship with prognosis. In addition, studies of the signal pathways involved in the lncRNA SNHG16–miR183-5p–FOXO1 axis can also be carried out to promote development of the pathogenesis network of AML. Interestingly, Zheng et al reported that miR183-5p has opposite effects on leukemia cell lines.26 THP1 cells are employed as the research object in this study, while HL60 cells and U937 cells were used in Zheng et al’s, which may be the reason for the opposite trend in our results. Regulation of the lncRNA SNHG16–miR183-5p–FOXO1 axis on HL60 cells and U937 cells will also be discussed in future studies.
Conclusion
This study discussed possible molecular mechanisms in AML via changes in cell characteristics caused by lncRNA SNHG16, miR183-5p, and FOXO1. lncRNA SNHG16 directly upregulated the promoting effect of FOXO1 on the proliferation of AML cells by coupling with miR183-5p, thus aggravating AML. Therefore, targeted regulation of the lncRNA SNHG16–miR183-5p–FOXO1 axis improve the condition of the disease.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM. Epidemiology of acute myeloid leukemia: recent progress and enduring challenges. Blood Rev. 2019;36:70–87. doi:10.1016/j.blre.2019.04.005
2. Kamath GR, Tremblay D, Coltoff A, et al. Comparing the epidemiology, clinical characteristics and prognostic factors of acute myeloid leukemia with and without acute promyelocytic leukemia. Carcinogenesis. 2019;40(5):651–660. doi:10.1093/carcin/bgz014
3. Panagopoulou P, Skalkidou A, Marcotte E, et al.; group F, group N-S. Parental age and the risk of childhood acute myeloid leukemia: results from the childhood leukemia international consortium. Cancer Epidemiol. 2019;59:158–165. doi:10.1016/j.canep.2019.01.022
4. Karalexi MA, Dessypris N, Clavel J, et al.; group N-S. Coffee and tea consumption during pregnancy and risk of childhood acute myeloid leukemia: a childhood leukemia international consortium (CLIC) study. Cancer Epidemiol. 2019;62:101581. doi:10.1016/j.canep.2019.101581
5. Bassig BA, Engel LS, Langseth H, et al. Pre-diagnostic serum concentrations of organochlorines and risk of acute myeloid leukemia: a nested case-control study in the Norwegian Janus Serum Bank Cohort. Environ Int. 2019;125:229–235. doi:10.1016/j.envint.2019.01.066
6. Allegra A, Spatari G, Mattioli S, et al. Formaldehyde exposure and acute myeloid leukemia: a review of the literature. Medicina (Kaunas). 2019;55(10):638. doi:10.3390/medicina55100638
7. Liu S, Zhang W, Liu K, Liu Y. LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p. Biomed Pharmacother. 2019;114:108862. doi:10.1016/j.biopha.2019.108862
8. Wang X, Kan J, Han J, Zhang W, Bai L, Wu H. LncRNA SNHG16 functions as an oncogene by sponging MiR-135a and promotes JAK2/STAT3 signal pathway in gastric cancer. J Cancer. 2019;10(4):1013–1022. doi:10.7150/jca.29527
9. Lin Q, Zheng H, Xu J, Zhang F, Pan H. LncRNA SNHG16 aggravates tumorigenesis and development of hepatocellular carcinoma by sponging miR-4500 and targeting STAT3. J Cell Biochem. 2019;120:11604–11615. doi:10.1002/jcb.28440
10. Han W, Du X, Liu M, Wang J, Sun L, Li Y. Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int J Biol Macromol. 2019;121:270–278. doi:10.1016/j.ijbiomac.2018.10.004
11. Yang T, Jin X, Lan J, Wang W. Long non-coding RNA SNHG16 has tumor suppressing effect in acute lymphoblastic leukemia by inverse interaction on hsa-miR-124-3p. IUBMB Life. 2019;71(1):134–142. doi:10.1002/iub.1947
12. Zheng S, Zhong YF, Tan DM, Xu Y, Chen HX, Wang D. miR-183-5p enhances the radioresistance of colorectal cancer by directly targeting ATG5. J Biosci. 2019;44(4):92. doi:10.1007/s12038-019-9918-y
13. Lin D, Cui B, Ma J, Ren J. MiR-183-5p protects rat hearts against myocardial ischemia/reperfusion injury through targeting VDAC1. Biofactors. 2020;46:83–93. doi:10.1002/biof.1571
14. Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ, Wu WB. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol. 2019;70:42–54 e43. doi:10.1016/j.exphem.2018.10.011
15. Cheng Y, Xiang G, Meng Y, Dong R. MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod Biol. 2016;16(3):225–233. doi:10.1016/j.repbio.2016.07.002
16. Yan H, Sun BM, Zhang YY, et al. Upregulation of miR-183-5p is responsible for the promotion of apoptosis and inhibition of the epithelial-mesenchymal transition, proliferation, invasion and migration of human endometrial cancer cells by downregulating Ezrin. Int J Mol Med. 2018;42(5):2469–2480. doi:10.3892/ijmm.2018.3853
17. Meng F, Zhang L. miR-183-5p functions as a tumor suppressor in lung cancer through PIK3CA inhibition. Exp Cell Res. 2019;374(2):315–322. doi:10.1016/j.yexcr.2018.12.003
18. Li W, Cui X, Qi A, Yan L, Wang T, Li B. miR-183-5p acts as a potential prognostic biomarker in gastric cancer and regulates cell functions by modulating EEF2. Pathol Res Pract. 2019;215(11):152636. doi:10.1016/j.prp.2019.152636
19. Xing YQ, Li A, Yang Y, Li XX, Zhang LN, Guo HC. The regulation of FOXO1 and its role in disease progression. Life Sci. 2018;193:124–131. doi:10.1016/j.lfs.2017.11.030
20. Liu H, Ni Z, Shi L, Ma L, Zhao J. MiR-486-5p inhibits the proliferation of leukemia cells and induces apoptosis through targeting FOXO1. Mol Cell Probes. 2019;44:37–43. doi:10.1016/j.mcp.2019.02.001
21. Seedhouse CH, Mills KI, Ahluwalia S, et al. Distinct poor prognostic subgroups of acute myeloid leukaemia, FLT3-ITD and P-glycoprotein-positive, have contrasting levels of FOXO1. Leuk Res. 2014;38(1):131–137. doi:10.1016/j.leukres.2013.10.030
22. Majumder S, Ren L, Pushpakumar S, Sen U. Hydrogen sulphide mitigates homocysteine-induced apoptosis and matrix remodelling in mesangial cells through Akt/FOXO1 signalling cascade. Cell Signal. 2019;61:66–77. doi:10.1016/j.cellsig.2019.05.003
23. Deng C, Zhong Z, Wu D, et al. Role of FoxO1 and apoptosis in pulmonary vascular remolding in a rat model of chronic thromboembolic pulmonary hypertension. Sci Rep. 2017;7(1):2270. doi:10.1038/s41598-017-02007-5
24. Huang C, Chen D, Zhu H, Lv S, Li Q, Li G. LITAF enhances radiosensitivity of human glioma cells via the FoxO1 pathway. Cell Mol Neurobiol. 2019;39(6):871–882. doi:10.1007/s10571-019-00686-4
25. Kode A, Mosialou I, Manavalan SJ, et al. FoxO1-dependent induction of acute myeloid leukemia by osteoblasts in mice. Leukemia. 2016;30(1):1–13. doi:10.1038/leu.2015.161
26. Zheng Z, Zheng X, Zhu Y, et al. miR-183-5p inhibits occurrence and progression of acute myeloid leukemia via targeting erbin. Mol Ther. 2019;27(3):542–558. doi:10.1016/j.ymthe.2019.01.016
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.