Back to Journals » Cancer Management and Research » Volume 12
lncRNA SNHG1 Promotes Progression of Cervical Cancer Through miR-195/NEK2 Axis
Received 22 August 2020
Accepted for publication 15 October 2020
Published 9 November 2020 Volume 2020:12 Pages 11423—11433
DOI https://doi.org/10.2147/CMAR.S277064
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Yuan Yuan Ji,1 Man Meng,2 Ye Miao3
1Department of Obstetrics and Gynecology, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, People’s Republic of China; 2Department of Geriatrics, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, People’s Republic of China; 3Department of Neurosurgery, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, People’s Republic of China
Correspondence: Ye Miao
Department of Neurosurgery, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou 121001, People’s Republic of China
Email [email protected]
Objective: Cervical cancer is a common gynecologic cancer, and no study has been reported on the way through which lncRNA SNHG1, miR-195 and NEK2 jointly affect cervical cancer cells (CCCs), so this paper will explore a new approach to the development of cervical cancer in this respect.
Methods: Altogether 72 cervical cancer tissues and 54 adjacent tissues were collected. qPCR was performed to quantify lncRNA SNHG1 and miR-195, whose expression vectors were constructed and then transfected into CCCs, so as to observe their effects on the cells. Western blotting (WB) was carried out to detect protein levels. MTT assay was conducted to detect cell activity. Flow cytometry was performed to detect cell apoptosis. Transwell was carried out to detect cell invasion and migration.
Results: The expression of lncRNA SNHG1 up-regulated while that of miR-195 down-regulated in CCCs. lncRNA SNHG1 regulated NEK2 through its targeted binding to miR-195. The down-regulation of lncRNA SNHG1 or the up-regulation of miR-195 led to the decrease of NEK2 and the reduction of cells’ activity, migration and invasion, also resulting in the increase of cell apoptosis. Rescue experiments showed that the down-regulation of miR-195 could offset the cell changes caused by lncRNA SNHG1.
Conclusion: lncRNA SNHG1 promotes the progression of cervical cancer through the miR-195/NEK2 axis, so lncRNA SNHG1, miR-195 and NEK2 may have potential values for diagnosing and treating cervical cancer.
Keywords: cervical cancer, lncRNA SNGH1, miR-195, NEK2
Introduction
Cervical cancer is a common female cancer.1 With respect to its pathological pathways, human papillomavirus (HPV) is a major cause,2,3 and changes in gene expression are also an inducing factor.4–6 Besides, marital status, drinking, smoking, chlamydia trachomatis and sexual hygiene are all risk factors for the disease.7–10 Currently, the pathogenic mechanism of cervical cancer is a research hotspot in the field of gynecologic cancers. Non-coding RNAs, such as long-stranded non-coding RNAs (lncRNAs) and microRNAs (miRNAs), play an important role in the development and progression of cervical cancer. Therefore, studying the relationship between the two RNAs and cervical cancer is beneficial to the treatment or early diagnosis of the disease.
Epigenetics is a regulatory mechanism of genes and does not involve their sequence changes. It is through this mechanism that non-coding RNAs that cannot participate in gene coding can change gene expression. lncRNAs bind to downstream sequences through specific sites, and thus regulate the expression of these sequences. As a lncRNA that is located on human chromosome 11, lncRNA SNHG1 has potential diagnostic and prognostic values, because it regulates the progression of cancers or diseases by binding to various miRNAs.11–15 This lncRNA is closely related to the development of cervical cancer, so its abnormal expression may lead to the cancerization of cervical cancer cells (CCCs).17 miRNAs and lncRNAs have similar mechanisms of action. miR-195, which is located on human chromosome 17, is a small RNA with a length of about 87 bp. Although it cannot participate in gene coding directly, it regulates the post-transcriptional levels of downstream mRNAs at the epigenetic level, and thereby affects gene translation. The abnormal expression of miR-195 is closely correlated with the development of breast cancer and other cancers.18–22 Moreover, previous studies have pointed out that the down-regulation of this miR is associated with the malignant cancerization of CCCs.23–25
In this paper, cervical cancer samples were detected, and we found that lncRNA SNHG1 up-regulated while miR-195 down-regulated in them. Meanwhile, it was predicted through the DIANA database that lncRNA SNHG1 had a targeted relationship with miR-195. NEK2 encodes a mitosis-related serine/threonine protein kinase. We found in this study that the down-regulation of lncRNA SNHG1 or the up-regulation of miR-195 could inhibit NEK2 in CCCs. Accordingly, it is speculated that lncRNA SNHG1, miR-195 and NEK2 may be related to the progression of cervical cancer. The way through which the three genes affect the development of cervical cancer has been rarely studied, so this paper has discussed a possible pathogenic mechanism of the disease.
Materials and Methods
Cervical Cancer Samples
Altogether 72 cervical cancer tissues and 54 normal adjacent tissues were collected in our hospital. Inclusion criteria were as follows: patients who were confirmed with cervical cancer according to clinical features or pathological sections; patients without mental diseases. Exclusion criteria were as follows: patients who were complicated with other tumors; patients who had been previously treated for cervical cancer; patients who did not cooperate in their treatment; patients complicated with other chronic gynecological diseases. The hospital informed the patients of the research information throughout the study, and all study participants provided written informed consents. This study has been approved by the First Affiliated Hospital of Jinzhou Medical University Ethics Committee. The tissues were sampled, sectioned, and then stored at −80°C for testing.
Cell Culture and Transfection
Human immortalized epidermal cells (HaCaT) and cervical cancer cell lines (HeLa/CasK/ME-180/C33A) were purchased from ATCC cell bank. The culture medium was composed of basal DMEM (Hyclone), 10% fetal bovine serum (FBS) solution (Gibco), and 1% penicillin/streptomycin solution (100X, Solarbio). The cells were inoculated into a T25 cell culture flask (Thermo Fisher), which was added with a culture medium (5 mL) that was preheated to 37°C. They were cultured (37°C, 5% CO2) in an animal cell incubator (Binder, German) until they grew well. The medium was replaced with an FBS-free medium before transfection, and the cells were inoculated into a 6-well plate at 1×105 cells/well during transfection. Purchased from Sangon Biotech (Shanghai) Co., Ltd., lncRNA SNHG1 siRNA, miR-195-mimics, miR-195-inhibitor and NC vectors were transfected into the cells using Lipofectamine 2000 transfection kits (Invitrogen, USA), with operating steps conducted according to the kit instruction. After 8-hour transfection, a fresh culture medium was exchanged to avoid poisoning cells.
RT-PCR
The tissue samples were ground to prepare into homogenates, and adherent cells were digested with trypsin to prepare into a cell suspension. The Trizol method was used to draw total RNA from the tissues or cells, and its optical density (OD) values at 260–280 nm were obtained using an ultraviolet spectrophotometer. The total RNA with OD260/OD280 >1.8 was used for subsequent RT-PCR quantification. Reverse transcription-fluorescence quantitative kits (TransGen, Beijing) and an ABI PRISM 7000 instrument (Applied Biosystems, USA) were used for the quantitative analysis of the RNA to be detected, using FastKing one-step method. Primers for lncRNA SNHG1, miR-195 and NEK2 were designed and synthesized by Sangon Biotech (Shanghai) Co., Ltd. The primer sequences are shown in Table 1. The reaction system of qPCR (50 μL) was as follows: 1.25 μL of upstream primers, 1.25 μL of downstream primers, 1.0 μL of probes, 10 pg/μg of RNA templates, 5 μL of 50×ROX Reference Dye ROX, and RNase-Free ddH2O that was finally added to make up to 50 μL. The reaction process was as follows: cycling (reverse transcription at 50°C for 30 min) for once; cycling (pre-denaturation at 95°C for 3 min) for once; cycling (denaturation at 95°C for 15 s and annealing at 60°C for 30 s) for 40 times. The results were analyzed by the ABI PRISM 7000 instrument. Internal reference genes were U6 and GAPDH, which were standardized by 2−ΔΔCt method.
 |
Table 1 Primer Sequences |
Western Blotting (WB)
20 mmol/L of Tris-HCl solution (pH7.5, Solarbio) was evenly mixed with a protein inhibitor (Solarbio) to form cell protein extracts. The adherent cells were digested with 1 mL of trypsin (Solarbio, 0.25×) to prepare into a suspension, which was added with 1 mL of the extracts and repeatedly blown until the cells completely lysed. The extracts were centrifuged at 1.6×104 ×g and 4°C in a precooling centrifuge for 20 min. The supernatant was taken out and the protein concentration was determined using BCA protein assay kits (Thermo Fisher). Next, the protein was separated with SDS-PAGE, then transferred to PVDF membrane (EMD millipore), and finally allowed to stand at room temperature for 1 hour (sealed with 5% skimmed milk-PBS solution). After that, the membrane was added with the protein to be tested and β-actin primary antibody, and then allowed to stand overnight at 4°C. After washed with PBS solution for 3 times, it was added with goat anti-rabbit secondary antibody (HRP cross-linked), and then allowed to stand at room temperature for 1 hour. Finally, it was washed with PBS solution, and ECL luminescent solution was used for visualization. With β-actin used as an internal reference, the relative expression level of the protein to be detected = the gray value of the band to be detected/the gray value of the β-actin band. NEK2, apoptosis-related proteins (Caspase-3/Caspase-9/Bax/Bcl-2), migration- and invasion-related proteins (E-cadherin/N-cadherin/β-catenin), β-actin and goat anti-rabbit (HRP cross-linked) were all purchased from Abcam.
Transwell
The cells were inoculated into the upper chamber of migration at 2×104 cells/well (the upper chamber was added with 200 μL of the mixed solution containing 10% FBS and 1% DMEM in advance), and 500 μL of DMEM (containing 10% FBS) was added to the lower chamber. Both chambers were cultured at 37°C and with 5% CO2 for 24 hours. Next, after liquid in the upper chamber was removed and the adherent cells were wiped off, the cells on the opposite side of the Transwell chamber were fixed with 4% paraformaldehyde for 20 min, and then stained with crystal violet for 15 min. Finally, PBS buffer solution was used to clean the chamber. The photographs of cell migration were collected under a 200-fold microscope, and cell number was calculated in 3 visual fields that were randomly selected, with the average value considered as the number of cells penetrating the membrane. The experiment was repeatedly conducted for 3 times. As for experiments for cell invasion, 8% matrigel was paved based on the above steps, and the number of cells in each well was increased to 5×104 cells/well.
MTT Assay
MTT (Sigma-Aldrich) was dissolved in DMSO (Solarbio) at a concentration of 5 mg/mL. Four 96-well plates were taken out, and 3 wells in each plate were randomly selected for inoculation at a density of 5×103 cells/100 μL per well. These plates were respectively taken out at 24, 48, 72 and 96 hours after inoculation, and then added with MTT solution at 10 μL/well, and finally cultured at 37°C and with 5% CO2 for 4 hours. The culture medium was removed, and the OD values at 570 nm were measured using a microplate reader.
Flow Cytometry
The cells lysed with trypsin to prepare into a cell suspension in which their number was controlled to 1×106. They were fixed with 70% ice-cold ethanol at 4°C for 30 min. After the ethanol was removed, the mixed solution of propidium iodide (50 ng/mL)/RNase (0.2 mg/mL)/0.1% Triton X-100 was added to the cells, which were then incubated at room temperature for 30 min. The FACScan flow cytometer (Becton Dickinson, USA) was used to analyze the cell apoptosis.
Dual-Luciferase Reporter Gene Assay
The DIANA database was used to predict the target sites of miR-195 and lncRNA SNHG1. The cells were inoculated into a 24-well plate (5×103 cells/well) and cultured until they grew well. The vectors of GLO-SNHG1-wt (containing miR-195 binding site) and GLO-SNHG1-mut (without miR-195 binding site) were constructed, and co-transfected with miR-195-mimics and NC-mimics into CCCs, respectively. After 48-hour transfection, luciferase intensities were detected using the dual-luciferase reporter gene assay system (Promega).
Statistics and Analysis
The data were statistically analyzed using SPSS 20.0 (Asia Analytics Formerly SPSS China), and the figures were plotted using Graphpad 8.0. Measurement data were expressed as mean±SD. Their comparison between two groups was conducted by an independent samples t test, and the comparison between multiple groups was conducted by one-way analysis of variance, and LSD-t test was used for post hoc pairwise comparison. All data were analyzed by a two-tailed test. 95% was considered as the confidential interval, and the difference was statistically significant when P<0.05.
Results
Up-Regulation of lncRNA SNHG1 and Down-Regulation of miR-195
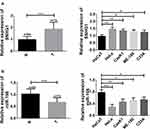
Altogether 72 cervical cancer tissues and 54 normal adjacent tissues were collected, in which the levels of lncRNA SNHG1 and miR-195 were quantified by RT-PCR. Figure 1 shows increasing lncRNA SNHG1 and decreasing miR-195 in cervical cancer tissues compared with those in normal adjacent tissues. Then, the levels of the two genes in CCCs and human cervical epithelial cells were detected by RT-PCR. The results showed up-regulating lncRNA SNHG1 and down-regulating miR-195 in both cells. This indicates that lncRNA SNHG1 up-regulates and miR-195 down-regulates in cervical cancer and that the changes may involve the malignant biological behaviors of CCCs. Thus, this paper focused on the relationship between the two genes and cervical cancer. Additionally, lncRNA SNHG1 has the highest expression in HeLa cells, which were therefore selected as the research objects for this study.
Targeted Relationship of lncRNA SNHG1/miR-195/NEK2 Axis
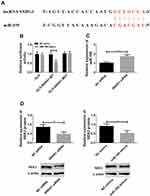
lncRNA SNHG1 and miR-195 showed an expression trend of “one high and one low” in cervical cancer, so it was speculated that lncRNA SNHG1 may bind to miR-195 through a specific site and thus lower its level. The DIANA database was used to predict and find the binding site between the two genes, and the results showed that there was a sequence fragment that could pair with and bind to miR-195 in the 3ʹ non-coding region of lncRNA SNHG1 (Figure 2A). The dual-luciferase reporter gene assay was performed to verify the existence of this site, and the results showed that the luciferase activity decreased after miR-195-mimics and lncRNA SNHG1 wt (Figure 2B) were co-transfected into the cells. Figure 2C and D showed that decreasing lncRNA SNHG1 increased miR-195. This indicates that lncRNA SNHG1 can bind to miR-195 and thus inhibit its level. Interestingly, the down-regulation of lncRNA SNHG1 or the up-regulation of miR-195-mimics can down-regulate NEK2, which reveals that NEK2 may be a downstream target gene of miR-195-mimics.
lncRNA SNHG1 Promoted Hyperproliferation and Malignant Proliferation of CCCs
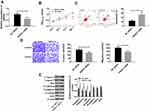
In order to explore the relationship between the up-regulation of lncRNA SNHG1 and cervical cancer, lncRNA siRNA vectors were constructed to down-regulate lncRNA SNHG1 (Figure 3A). MTT assay was performed to detect cell activity. Transwell was carried out to detect cell migration and invasion. Flow cytometry was performed to detect the apoptotic rate of cells. WB was performed to detect the levels of related proteins. Figure 3B shows that the down-regulation of lncRNA SNHG1 decreased cell activity and inhibited cell proliferation. Figure 3C shows that the down-regulation induced an increase in the apoptotic rate. Caspase-3, Caspase-9 and Bax are pro-apoptotic proteins, while Bcl-2 is an anti-apoptotic protein. During cell apoptosis, the pro-apoptotic proteins increased while the anti-apoptotic protein decreased. E-cadherin, N-cadherin and β-catenin are proteins related to cell migration and invasion. E-cadherin can inhibit cell invasion, while N-cadherin and β-catenin can promote cell migration and invasion. Figure 3D shows that the down-regulation reduced the migrating and invasive cells, and inhibited cell invasion and migration. Figure 3C and D showed that the down-regulation decreased cell migration and invasion but increased cell apoptosis. Therefore, the detection of related proteins can help understand the influences of lncRNA SNHG1 on cell apoptosis, invasion and migration from the perspective of molecular biology. Figure 3E shows that the down-regulation decreased N-cadherin, β-catenin and Bcl-2, but increased Caspase-3, Caspase-9, Bax and E-cadherin. These findings reveal that lncRNA SNHG1 promotes the hyperproliferation and malignant proliferation of CCCs, so the down-regulation of this gene inhibits the hyperproliferation and malignant proliferation and promotes cell apoptosis.
miR-195 Inhibited Hyperproliferation and Malignant Proliferation of CCCs
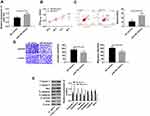
Since miR-195 is regulated by lncRNA SNHG1 and down-regulates in cervical cancer, its role in the disease was investigated in this study. miR-195-mimics vectors were constructed to up-regulate miR-195 (Figure 4) and the effects on CCCs were observed. The results showed that the up-regulation of miR-195 decreased cell activity, as well as inhibited cell migration and invasion and promoted cell apoptosis through down-regulating N-cadherin, β-catenin and Bcl-2 and up-regulating Caspase-3, Caspase-9, Bax and E-cadherin. These findings indicate that miR-195 inhibits the hyperproliferation and malignant proliferation of CCCs.
Rescue Experiments
Figure 5A and B showed that lncRNA SNGH1 regulated NEK2 through miR-195. Figure 5C and D showed that lncRNA SNHG1 and miR-195 regulated the biological behaviors of CCCs. Therefore, it was speculated that the two genes may regulate the behaviors through NEK2. In order to verify this inference, lncRNA SNHG1 siRNA and LNC RNA SNHG1 siRNA+miR-195-inhibitor were respectively transfected into CCCs, so as to observe their changes. Figure 5E shows that the down-regulation of miR-195 could offset the decrease of NEK2, the reduction of cells’ activity, migration and invasion, and the increase of cell apoptosis, which were caused by the down-regulation of lncRNA SNHG1. These findings indicate that lncRNA SNHG1 promotes the hyperproliferation and malignant proliferation of CCCs through the miR-195/NEK2 axis.
Discussion
As a malignant gynecological tumor that occurs in the cervix, cervical cancer is difficult to precisely diagnose owing to its unapparent early features. At present, the treatment and diagnosis of the disease mainly depend on patients’ regular inspection and self-management awareness due to the high occult nature of it. Therefore, how to effectively prevent and treat cervical cancer is the main purpose of research on the disease. Non-coding RNAs have attracted researchers’ attention due to their unique regulatory mechanisms and potential diagnostic/therapeutic values. During the detection of cervical cancer samples, lncRNA SNHG1 was found to up-regulate and miR-195 was found to down-regulate. Their abnormal expression seemed to be related to the development and progression of cervical cancer. lncRNAs regulating miRNAs is a common molecular regulatory mechanism in cells, so when the two RNAs have abnormal expression, the cell regulation in which they are involved will be inevitably out of control. In addition, the regulation of their levels in CCCs resulted in changes in NEK2 expression. The lncRNA/miRNA axis can also affect the normal functioning of cells via binding to downstream target genes, so the abnormal expression of miR-195 and lncRNA SNHG1 may be the main reason for the changes in NEK2. Therefore, it is speculated that lncRNA SNHG1, miR-195 and NEK2 may regulate the development of cervical cancer through some connection.
lncRNA SNHG1 and miR-195 showed an expression trend of “one high and one low” in cervical cancer (Figure 1), which was similar to others’ findings.17,23–25 Therefore, it is speculated that lncRNA SNHG1 may bind to miR-195 through a specific site and thus lower its level. The DIANA database was used to predict and find the binding site between the two genes, and the results showed that there was a sequence fragment that could pair with and bind to miR-195 in the 3ʹ non-coding region of lncRNA SNHG1 (Figure 2A). The dual-luciferase reporter gene assay was performed to verify the existence of this site, and the results showed that the luciferase activity decreased when miR-195-mimics and lncRNA SNHG1 wt were co-transfected into the cells (Figure 2B). This indicates that lncRNA SNHG1 can bind to miR-195 and thus inhibit its level. Interestingly, the down-regulation of lncRNA SNHG1 or the up-regulation of miR-195-mimics can down-regulate NEK2. Accordingly, lncRNA SNHG1 may regulate NEK2 through binding to miR-195, thus participating in the progression of cervical cancer.
In this paper, corresponding expression vectors were constructed to explore the specific effects of lncRNA SNHG1, miR-195 and NEK2 on cervical cancer. WB was conducted to detect the levels of NEK2, Caspase-3, Caspase-9, Bax, Bcl-2, E-cadherin, N-cadherin and β-catenin. MTT assay was performed to detect cell activity. Flow cytometry was carried out to detect cell apoptosis. Transwell was conducted to detect cell invasion and migration. Figures 3 and 4 show that the down-regulation of lncRNA SNHG1 or the up-regulation of miR-195 decreased cell activity and NEK2, increased cell apoptosis, and reduced cell malignant proliferation; the down-regulation or the up-regulation also decreased N-cadherin, β-catenin and Bcl-2, but increased Caspase-3, Caspase-9, Bax and E-cadherin. Caspase-3, Caspase-9, Bax, E-cadherin, Bcl-2, N-cadherin and β-catenin are proteins related to cell proliferation, apoptosis, invasion and migration, so detecting their levels can help know the mechanism of changes in the above-mentioned biological functions. The rescue experiments showed that the down-regulation of miR-195 can offset the increase, the decrease and the reduction, which were caused by the down-regulation of lncRNA SNHG1 (Figure 5). During cell mitosis, NEK2 protein not only promotes centriole separation, but also monitors correct chromosome division.26 Passive or active changes in its level are very likely to lead to the disorders of cell life process. Previous studies believe that highly active NEK2 protein is positively correlated with the development of cervical cancer.26,27 Therefore, in this paper, the down-regulation of NEK2, which was caused by lncRNA SNHG1 down-regulation and miR-195 up-regulation, leads to the disorders of cell mitosis and thus inhibits cell activity and proliferation. However, the down-regulation of miR-195 offset the down-regulation of NEK2 that was caused by lncRNA SNHG1 down-regulation, which therefore restored cell mitosis and enhanced cell activity and proliferation. These findings confirm that lncRNA SNHG1 up-regulates NEK2 by down-regulating miR-195, thereby promoting the up-regulation of cervical cancer.
In this paper, cell experiments showed a close relationship between the lncRNA SNHG1/miR-195/NEK2 axis and cervical cancer. miR-195 and NEK2 might have negative regulation, but the targeted relationship between miR-195 and NEK2 was not explored in this paper, which is the limitation. Therefore, their targeted sites will be found and confirmed in the following research, and whether the axis has potential values for diagnosing and treating cervical cancer will be discussed, so as to provide reliable and scientific data for promoting the research on the disease. In addition, it is essential to study whether the three genes affect the resistance of CCCs to anti-cancer drugs (such as cisplatin and paclitaxel), so subsequent research may be beneficial to the development of the target therapy for cervical cancer.
Conclusion
In summary, lncRNA SNHG1 promotes the effects of NEK2 on CCCs through down-regulating miR-195, thus inducing the development and progression of cervical cancer. Therefore, lncRNA SNHG1, miR-195 and NEK2 may help to improve the diagnostic efficiency and therapeutic effect on the disease.
Funding
This study was supported by 2017 Youth Fund Project No. of Liaoning Provincial Department of education (Grant No.: jytqn201708).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi:10.1016/S0140-6736(18)32470-X
2. Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8:e191–e203. doi:10.1016/S2214-109X(19)30482-6
3. Okunade KS. Human papillomavirus and cervical cancer. J Obstet Gynaecol. 2019;1–7. doi:10.1080/01443615.2019.1634030
4. Du GH, Wang JK, Richards JR, Wang JJ. Genetic polymorphisms in tumor necrosis factor alpha and interleukin-10 are associated with an increased risk of cervical cancer. Int Immunopharmacol. 2019;66:154–161. doi:10.1016/j.intimp.2018.11.015
5. De Strooper LMA, Berkhof J, Steenbergen RDM, et al. Cervical cancer risk in HPV-positive women after a negative FAM19A4/mir124-2 methylation test: a post hoc analysis in the POBASCAM trial with 14 year follow-up. Int J Cancer. 2018;143:1541–1548. doi:10.1002/ijc.31539
6. Wang Y, Luo T. LINC00673 rs11655237 polymorphism is associated with increased risk of cervical cancer in a Chinese population. Cancer Control. 2018;25(1):1073274818803942. doi:10.1177/1073274818803942
7. Sharma P, Pattanshetty SM. A study on risk factors of cervical cancer among patients attending a tertiary care hospital: a case-control study. Clin Epidemiol Glob Health. 2018;6:83–87. doi:10.1016/j.cegh.2017.10.001
8. Sugawara Y, Tsuji I, Mizoue T, et al. Cigarette smoking and cervical cancer risk: an evaluation based on a systematic review and meta-analysis among Japanese women. Jpn J Clin Oncol. 2019;49:77–86. doi:10.1093/jjco/hyy158
9. Kashyap N, Krishnan N, Kaur S, Ghai S. Risk factors of cervical cancer: a case-control study. Asia Pac J Oncol Nurs. 2019;6(3):308–314. doi:10.4103/apjon.apjon_73_18
10. Zhu H, Shen Z, Luo H, Zhang W, Zhu X. Chlamydia trachomatis infection-associated risk of cervical cancer: a meta-analysis. Medicine. 2016;95:e3077. doi:10.1097/MD.0000000000003077
11. Liu T, Zuo JJ, Li F, Xu YC, Zheng AY, Tao ZZ. LncRNA SNHG1 promotes cell proliferation in laryngeal cancer via Notch1 signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:6562–6569.
12. Li SJ, Wang L, Sun ZX, Sun SJ, Gao J, Ma RL. LncRNA SNHG1 promotes liver cancer development through inhibiting p53 expression via binding to DNMT1. Eur Rev Med Pharmacol Sci. 2019;23:2768–2776.
13. Li X, Zheng H. LncRNA SNHG1 influences cell proliferation, migration, invasion, and apoptosis of non-small cell lung cancer cells via the miR-361-3p/FRAT1 axis. Thorac Cancer. 2020;11:295–304. doi:10.1111/1759-7714.13256
14. Lan X, Liu X. LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med. 2019;23:2351–2361. doi:10.1111/jcmm.13497
15. Zhang N, Liu FL, Ma TS, Zhang ZZJ. LncRNA SNHG1 contributes to tumorigenesis and mechanism by targeting miR-338-3p to regulate PLK4 in human neuroblastoma. Eur Rev Med Pharmacol Sci. 2019;23:8971–8983.
16. Thin KZ, Tu JC, Raveendran S. Long non-coding SNHG1 in cancer. Clin Chim Acta. 2019;494:38–47. doi:10.1016/j.cca.2019.03.002
17. Liu Y, Yang Y, Li L, et al. LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol. 2018;96:38–43. doi:10.1139/bcb-2017-0188
18. McAnena P, Tanriverdi K, Curran C, et al. Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer. 2019;19(1):436. doi:10.1186/s12885-019-5636-y
19. Purohit PK, Edwards R, Tokatlidis K, Saini N. MiR-195 regulates mitochondrial function by targeting mitofusin-2 in breast cancer cells. RNA Biol. 2019;16:918–929. doi:10.1080/15476286.2019.1600999
20. Maroof H, Irani S, Arianna A, Vider J, Gopalan V, Lam AK. Interactions of vascular endothelial growth factor and p53 with miR-195 in thyroid carcinoma: possible therapeutic targets in aggressive thyroid cancers. Curr Cancer Drug Targets. 2019;19:561–570. doi:10.2174/1568009618666180628154727
21. Yu X, Zhang Y, Wu B, Kurie JM, Pertsemlidis A. The miR-195 axis regulates chemoresistance through TUBB and lung cancer progression through BIRC5. Mol Ther Oncolytics. 2019;14:288–298. doi:10.1016/j.omto.2019.07.004
22. Chae DK, Park J, Cho M, et al. MiR-195 and miR-497 suppress tumorigenesis in lung cancer by inhibiting SMURF2-induced TGF-beta receptor I ubiquitination. Mol Oncol. 2019;13:2663–2678. doi:10.1002/1878-0261.12581
23. Zhou Q, Han LR, Zhou YX, Li Y. MiR-195 suppresses cervical cancer migration and invasion through targeting Smad3. Int J Gynecol Cancer. 2016;26:817–824. doi:10.1097/IGC.0000000000000686
24. Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25:637–644. doi:10.1080/1061186X.2017.1307379
25. Li Z, Wang H, Wang Z, Cai H. MiR-195 inhibits the proliferation of human cervical cancer cells by directly targeting cyclin D1. Tumour Biol. 2016;37:6457–6463. doi:10.1007/s13277-015-4540-6
26. Naro C, Barbagallo F, Chieffi P, Bourgeois CF, Paronetto MP, Sette C. The centrosomal kinase NEK2 is a novel splicing factor kinase involved in cell survival. Nucleic Acids Res. 2014;42:3218–3227.
27. Koch M, Wiese M. Gene expression signatures of angiocidin and darapladib treatment connect to therapy options in cervical cancer. J Cancer Res Clin Oncol. 2013;139:259–267. doi:10.1007/s00432-012-1317-9
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.