Back to Journals » OncoTargets and Therapy » Volume 12
lncRNA small nucleolar RNA host gene 20 predicts poor prognosis in glioma and promotes cell proliferation by silencing P21
Authors Li XS, Shen FZ, Huang LY, Hui L, Liu RH, Ma YJ, Jin BZ
Received 29 October 2018
Accepted for publication 3 January 2019
Published 24 January 2019 Volume 2019:12 Pages 805—814
DOI https://doi.org/10.2147/OTT.S192641
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Xiang-Sheng Li,1 Fa-Zheng Shen,1 Li-Yong Huang,1 Lei Hui,1 Rui-Hua Liu,1 Yan-Juan Ma,2 Bao-Zhe Jin1
1Department of Neurosurgery, The First Affiliated Hospital of Xinxiang Medical University, Xinxiang 453000, Henan Province, People’s Republic of China; 2Department of Emergency, The First Affiliated Hospital of Xinxiang Medical University, Xinxiang 453000, Henan Province, People’s Republic of China
Background: In multiple cancers, long non-coding RNA small nucleolar RNA host gene 20 (lncRNA SNHG20) is generally dysregulated. In the present study, both the biological role and clinicopathological value of lncRNA SNHG20 in glioma are explored.
Methods: Real-time PCR was employed to determine lncRNA SNHG20 expression in glioma patients. The prognostic role of expression of lncRNA SNHG20 was evaluated in a retrospective cohort study. In addition, the association between lncRNA SNHG20 expression and the clinicopathological features of glioma patients, such as tumor recurrence, survival status, follow-up time, WHO grade, resection extent, tumor location, Karnofsky performance scale score, cystic change, tumor size, gender and age, was discussed. By constructing and transfecting siRNAs that targeted lncRNA SNHG20 into the glioma U87 cells, the effects of lncRNA SNHG20 on the proliferation and cell cycle of U87 cells were assessed through cell counting kit-8, colony formation and cell cycle assays, respectively. In addition, Western blot and real-time PCR measured the expression levels of P21 and CCNA1 in U87 cells after being transfected with SNHG20 siRNA.
Results: Our results suggested the high expression of lncRNA SNHG20 in human glioma tissues compared with normal brain tissues, which was related to recurrence-free survival and poor overall survival in glioma patients. According to the existing retrospective cohort study, high lncRNA SNHG20 expression was associated with tumor size, extent of resection, WHO grade, follow-up time, survival status and recurrence. Besides, knocking down the expression of lncRNA SNHG20 could inhibit the proliferation and colony formation abilities of glioma U87 cells through cell cycle arrest. Consequently, the expression of CCNA1 was inhibited, and the expression of P21 was up-regulated in U87 cells.
Conclusion: A high lncRNA SNHG20 expression level predicts the poor prognosis for glioma patients. Moreover, lncRNA SNHG20 can promote glioma proliferation through silencing P21 and thus lncRNA SNHG20 is an independent potential prognostic biomarker for glioma patients.
Keywords: lncRNA SNHG20, glioma, clinicopathological, prognosis, proliferation
Introduction
As the most common central nervous system malignant tumor, glioma has the characteristics of poor prognosis, aggressiveness, rapid progression and frequent recurrence.1 In particular, the median survival time of patients with glioblastoma was shorter than 1 year.2 Obviously, dismal was remained in the prognosis for glioma patients although there are a variety of treatment options, such as chemotherapy, radiotherapy and surgery.3 So far, few useful biomarkers have been used to monitor the burden and response to treatment of gliomas. Therefore, prognostic evaluation and early diagnosis of glioma are essential for early treatment and improvement of survival rate.
Small nucleolar RNA host gene 20 (SNHG20) was originally identified in hepatocellular carcinoma through microarray data, which was a recently identified lncRNA and located at chromosome 17q25.2 position in the human genome.4 Recent studies demonstrated that lncRNA SNHG20 was dysregulated in multiple cancer and it exerts an important role in tumor growth, metastasis, invasion and poor survival.5 As a result, lncRNA SNHG20 is of great importance to tumor progression. However, the role of lncRNA SNHG20 in the prognosis and tumorigenesis of gliomas has not been fully clarified.
Admittedly, the function and prognostic value of lncRNA SNHG20 in glioma were first examined in the present study. The study results indicated the up-regulation of lncRNA SNHG20 expression in glioma tissues. Subsequently, the function and clinical significance of lncRNA SNHG20 expression in human glioma were explored.
Patients and methods
Patients and tissue samples
One hundred eight glioma patients undergoing an initial surgery at the First Affiliated Hospital of Xinxiang Medical University were enrolled into this study from 2011 to 2017. Additionally, epileptic resections obtained 12 normal brain tissues (NBTs). All glioma patients were diagnosed in the pathology department. All samples were stored and frozen in liquid nitrogen. Besides, all patients were naive to therapy before resection. The informed consent was provided by each patient, and the research samples obtained the approval of the Medical Ethics Committee of the First Affiliated Hospital of Xinxiang Medical University.
Quantitative real-time polymerase chain reaction (qRT-PCR) assay
The Trizol reagent was used to extract the total RNA from cells and glioma tissues. qRT-PCR detected the expression of lncRNA SNHG20 mRNA through the one-step RT-PCR kit (Takara Bio, Inc., Otsu, Japan) according to the manufacturer’s protocol. The primers were obtained from Genechem Co. Ltd (Invitrogen, Shanghai, People’s Republic of China), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was the internal control. The primers included lncRNA SNHG20: forward, 5′-ATGGCTATAAATAGATACACGC-3′, and reverse, 5′-GGTACAAACAGGGAGGGA-3′; CCNA1: forward, 5′-ATTCATTAAGTGAAATTGTGC-3′; and reverse, 5′-CTTCCATTCAGAAACTTATTG-3′; P21: forward, 5′-CAGAGGAGGCGCCATGT-3′; and reverse, 5′-GGAAGGTAGAGCTTGGGCAG-3′; GAPDH: forward, 5′-GGAGTCAACGGATTTGGTCGTAT-3′ and reverse, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. The expression levels of lncRNA SNHG20 mRNA, P21 and CCNA1 were quantified by the 2−ΔΔCT method and further normalized by the expression level of GAPDH mRNA.
Cell lines and transfection
Human glioma cell line U87 cells purchased from Shanghai Cell Bank (Shanghai, People’s Republic of China) were cultured in DMEM (Thermo Fisher Scientific, Carlsbad, CA, USA), which were supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA) with 5% CO2 at 37°C.
The negative control siRNA (NC siRNA) and siRNA of lncRNA SNHG20 were purchased from Shanghai Genechem Biotechnology Co., Ltd. The 6×105 U87 cells achieved 70% confluence were put in a 6-well plate and infected with NC siRNA or SNHG20 siRNA. The real-time PCR validated the efficiency of SNHG20 siRNA transfection.
Western blotting
The tissues and cells were obtained under the instructions of the manufacturer, and a BCA Protein Assay Kit (Pierce, Rockford, IL, USA) was used to determine the protein concentration. The same amount of total protein was isolated by 12.5% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane. The first antibody was purchased from ABCAM (Cambridge, UK) Trading Co., Ltd., including rabbit anti-P21, anti-CCNA1 and anti-GAPDH antibodies, while horseradish peroxidase combined goat anti rabbit IgG (purchased from Boster Biological Technology Co. Ltd, Wuhan, People’s Republic of China) as secondary antibody. At the same time, GAPDH was used as internal control to normalize the expression level of lncRNA SNHG20.
Cell counting kit-8 (CCK-8) assay
U87 cells at exponential phase were collected, infected with SNHG20 siRNA or NC siRNA, inoculated at a density of 3.5×103 cells/well, and cultured in the 96-well plates. After 0, 24, 48 and 72 hours, each well was added with 10 μL of CCK-8 solution. After 2 hours, a microplate reader (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was adopted to measure the absorbance at 450 nm. Besides, a total of five repeats/groups were performed, three times independently.
Colony formation assay
After transfection with SNHG20 siRNA or NC siRNA, glioma U87 cells were counted, plated onto the 6-well cell culture plates at a density of 300 cells/well and incubated at 37°C for colony formation. Subsequently, the visible colonies were fixed with 4% paraformaldehyde for 30 minutes and stained with 0.1% crystal violet for another 30 minutes. Colony-forming efficiency was calculated as the number of colonies (with the diameter of >0.5 mm).
Flow cytometry
Glioma U87 cells were infected with SNHG20 siRNA or NC siRNA and incubated at 37°C before being harvested. Then, the cells were washed twice with PBS and subsequently fixed with 75% cold ethanol. The cells were treated with a cell cycle staining kit and incubated in darkness for 30 minutes as directed by the manufacturer. Eventually, the cells were analyzed through flow cytometry.
Statistical analysis
In the current study, the data were conducted with statistical analysis using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). The measurement data expressed as mean ± SD were analyzed through Student’s t-test between two groups. The chi-squared test was carried out to analyze the enumeration data. The Kaplan–Meier analysis was performed to evaluate the survival, and the survival rate was evaluated through the long-rank test. In addition, the Cox regression analysis was also employed to perform univariate and multivariate analyses of survival data. The P-value of <0.05 was considered as statistically significant.
Ethics approval and consent to participate
According to the Declaration of Helsinki of 1964 and all subsequent revisions, this study was reviewed and approved by the Ethics committee of the First Affiliated Hospital of Xinxiang Medical University and all patients provided written informed consent according to the local ethics committee regulations.
Results
lncRNA SNHG20 was upregulated in human glioma tissues
To determine the differential expression of lncRNA SNHG20 between NBTs and glioma tissues, the lncRNA SNHG20 expression levels were determined using real-time PCR assay in 108 glioma samples. The clinicopathological features of glioma patients were shown in Table 1. There was no significant difference in the age and gender distribution between the glioma and NBT groups. The results indicated that lncRNA SNHG20 mRNA levels were significantly higher than those in NBT (Figure 1). The results suggested that lncRNA SNHG20 may be involved in the development of glioma.
  | Figure 1 The lncRNA SNHG20 expression levels in glioma tissues are outstandingly higher than those in normal brain tissues. |
Association between lncRNA SNHG20 expression and clinicopathological characteristics of glioma
For statistical analysis, the glioma patients were classified into the high lncRNA SNHG20 expression group (n=54) and low lncRNA SNHG20 expression group (n=54) based on the median value of lncRNA SNHG20 expression level. Then, the association between lncRNA SNHG20 expression and clinicopathological characteristics in glioma patients was discussed. Table 2 demonstrates the association between the lncRNA SNHG20 and the tumor size (P=0.002), extent of resection (P=0.012), WHO grade (P=0.0031), follow-up time (P=0.000), survival status (P=0.000) and recurrence (P=0.000). However, the expression of lncRNA SNHG20 was not associated with tumor location, Karnofsky performance scale score, cystic change, gender and age (P>0.05). According to the Spearman analysis of the association between lncRNA SNHG20 and clinicopathological characteristics, the expression of lncRNA SNHG20 was associated with tumor size (P<0.002), extent of resection (P=0.011), WHO grade (P=0.031), follow-up time (P=0.000), survival status (P=0.000) and recurrence (P=0.000), as presented in Table 3.
  | Table 3 Spearman analysis of correlation between lncRNA SNHG20 and clinicopathological |
High lncRNA SNHG20 expression was associated with poor prognosis for glioma
To investigate the effect of lncRNA SNHG20 expression on the prognosis of gliomas, Kaplan–Meier analysis and log-rank test were performed in the current study. The results showed that the OS (Figure 2A, P<0.0001) in glioma patients with high expression were significantly lower than those in glioma patients with low expression. Kaplan–Meier survival analysis of lncRNA SNHG20 expression was performed in gliomas with different WHO grades and tumor sizes. According to the results, high expression of lncRNA SNHG20 indicated poor OS in both different grades (Figure 2B and C, I<0.01) and different tumor sizes (Figure 2D and E, P<0.01) glioma patients. Similar to previous results, high expression of lncRNA SNHG20 indicated poor RFS in total (Figure 2F, P<0.0001), different grades (Figure 2G and H, P<0.01) and different tumor sizes (Figure 2I and J, P<0.01) glioma patients.
All clinical data were conducted with multivariate and univariate Cox regression analyses. The results demonstrated that lncRNA SNHG20 expression was an independent prognostic factor for OS (Table 4) and RFS (Table 5) in patients with glioma (P<0. 05).
  | Table 4 Univariate and multivariate analyses for overall survival in glioma patients |
  | Table 5 Univariate and multivariate analyses for recurrence-free survival in glioma patients |
Inhibition of lncRNA SNHG20 expression by SNHG20 siRNA in human glioma U87 cells
SNHG20 siRNA and NC siRNA were transfected into human glioma U87 cells, so that the function of lncRNA SNHG20 in glioma can be explored. qRT-PCR was used to analyze the knockdown effects. After transfection, the lncRNA SNHG20 expression levels in U87 cells of SNHG20 siRNA group were lower than NC siRNA group (Figure 3).
Downregulation of lncRNA SNHG20 expression markedly inhibited glioma U87 cell proliferation and colony formation
CCK-8 assays were performed after glioma U87 cells transfected with SNHG20 siRNA or NC siRNA, and the effect of SNHG20 on glioma cell growth was examined. In Figure 4, the growth of U87 cells treated with SNHG20 siRNA was markedly inhibited compared to NC siRNA group. The glioma U87 cell growth was significantly reduced by SNHG20 siRNA cells on day 4 and day 5 in vitro. Similarly, after down-regulation of lncRNA SNHG20, colony formation was also dramatically reduced (P<0.01, Figure 5).
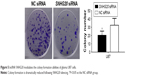  | Figure 5 lncRNA SNHG20 modulates the colony formation abilities of glioma U87 cells. |
Inhibition of lncRNA SNHG20 induces G0/G1 phase arrest in glioma U87 cells
Due to the direct association between proliferation and cell cycle distribution, we investigated the cell cycle distribution of U87 cells after down-regulation of lncRNA SNHG20. The cell cycle of U87 cells was detected by flow cytometry. As shown in Figure 6, the SNHG20 siRNA group displayed the following: (G0/G1 phase, 82.14%±5.32%; S phase, 11.94%±0.82%; and G2/M phase, 5.92%±0.64%), and the NC siRNA group displayed the following distribution: (G0/G1 phase, 58.27%±4.21%; S phase, 28.62%±2.36%; and G2/M phase, 13.11%±1.98%). With the absence of lncRNA SNHG20, the number of cells entering G0/G1 phase increased by 23.8%, and the number of cells entering S phase decreased by 16.7 cells (P<0.01). Knocking down lncRNA SNHG20 expression significantly increased the proportion of cells in G0/G1 phase and decreased the percentage of cells in S phase and G2/M phase. The above results suggest that lncRNA SNHG20 inhibits the proliferation of U87 cells through blocking the cell cycle progression in G0/G1 phase.
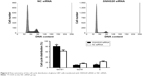  | Figure 6 Flow cytometry of the cell cycle distribution of glioma U87 cells transfected with SNHG20 siRNA or NC siRNA. |
The change of cell cycle-associated proteins after transfected with SNHG20 siRNA in human glioma U87 cells
As lncRNA SNHG20 affected cells proliferation through modulation of the cell cycle arrest, cell cycle-regulate gene expressions were examined at the transcriptional and translational levels. Furthermore, U87 cells were transfected with SNHG20 siRNA or NC siRNA, and the P21 and CCNA1 expression levels were determined by real-time PCR and Western blotting. The results showed that P21 mRNA and protein levels were significantly increased in glioma U87 cells transfected with SNHG20 siRNA compared to those transfected with NC siRNA. Furthermore, the results demonstrated that the CCNA1 protein levels were dramatically down-regulated after transfection with SNHG20 siRNA compared with that after transfection with NC siRNA (Figure 7).
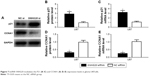  | Figure 7 lncRNA SNHG20 modulates the P21 (A–C) and CCNA1 (A, D, E) expression levels in glioma U87cells. |
Discussion
lncRNA lacking a meaningful open reading frame with various important functions in disease such as posttranscriptional, transcriptional and epigenetic regulation is defined as transcribed RNA molecules which is >200 nucleotides in length that is lack.6,7 In addition, lncRNA dysregulation is concerned with different types of cancer.8–11 For instance, some lncRNAs exert a crucial part in metastasis, invasion as well as cancer cell proliferation, showing that it might be a useful marker of cancer prognosis.12–14
Cancer still threats human health.15 The exact mechanism of metastasis remains uncertain in cancer patients despite the fact that metastasis is an important indicator of a poor prognosis.16,17 At present, new molecular markers should be identified to predict tumor metastasis due to its important role in treating and predicting cancer.18 lncRNA is one of the molecular markers which can influence the development and occurrence of tumors. It has the potential of collecting biomarkers which are beneficial for monitoring and diagnosing tumors.19
lncRNA SNHG20 is proved to be an important oncogene in various human cancers according to previous studies.20 According to the recent studies, it was confirmed that lncRNA SNHG20 expression is aberrantly high expression in ovarian cancer tissues and SNHG20 silencing could inhibit ovarian cancer progression through Wnt/β-catenin signaling pathway.21 Moreover, according to Wang et al, SNHG20 was significantly higher in osteosarcoma samples than that in the cancer-adjacent tissues, and high lncRNA SNHG20 promoted the osteosarcoma tumorigenesis through mitochondrial apoptosis pathway.22 Li et al23 reported that knockdown of SNHG20 inhibited colorectal cancer cell proliferation, cell cycle progression, migration and invasion, revealing that lncRNA SNHG20 might be a crucial prognostic factor for cancer patients. The function of lncRNA SNHG20 in prognosis and progression of glioma patients still remains unknown.
The present study aimed to investigate the clinical significance and role of lncRNA SNHG20 in glioma patients for the first time. First, our results showed that the expression of lncRNA SNHG20 in human gliomas was higher than that in normal brain tissues. Stastistical analysis demonstrated that the expression of lncRNA SNHG20 was closely related to tumor size, WHO grade, adjuvant therapy and recurrence. Third, Kaplan–Meier analysis proved that OS and RFS were shorter in glioma patients undergoing high expression of lncRNA SNHG20. In addition, further multivariate and univariate survival analyses confirmed that lncRNA SNHG20 could be used as an independent prognostic biomarker in glioma patients. SNHG20 siRNA was constructed and transfected to glioma U87 cells to detect the function of lncRNA SNHG20 in glioma cells, and the expression levels of lncRNA SNHG20 gene in U87 cells in vitro were down-regulated. The results indicated that after knockdown of lncRNA SNHG20 expression, the proliferation and colony formation abilities of glioma U87 cells were inhibited through increasing the percentage of cells at G0/G1 phase. To understand the potential molecular mechanisms, the potential target proteins involved in cell cycle progression were assessed. Here, CCNA1 expression levels decreased due to the loss of lncRNA SNHG20 in glioma U87 cells. There existed inverse association between lncRNA SNHG20 expression and P21 expression. CCNA1 alters cell cycle progression has been proved to induce carcinogenesis.24 P21 encoded by the CDKN1A gene on 6p21.2 in humans is a cyclin-dependent kinase inhibitor. P21 inhibits the complexes of CDK2 and CDK1 to mediate the p53-dependent cell cycle G1 phase arrest.23,25 In accordance with the results, lncRNA SNHG20 contributes to the proliferation of U87 cells via regulating CCNA1 and P21 expression. Nevertheless, the explicit mechanism of lncRNA SNHG20 should be evaluated by further validation and functional evaluation in other glioma cell lines.
Conclusion
To conclude, lncRNA SNHG20 is of great important to the progression and prognosis of glioma. Based on the high expression of lncRNA SNHG20 in human glioma tissues, the high lncRNA SNHG20 expression predicts dismal prognosis for glioma patients. Besides, lncRNA SNHG20 expression can be downregulated by SNHG20 siRNA, and the proliferation abilities of glioma U87 cells would be inhibited. Taken together, lncRNA SNHG20 can be taken as an independent prognostic biomarker for glioma.
Acknowledgment
This article was supported by the Youth Fund Project of the First Affiliated Hospital of Xinxiang Medical University (No QN-2017-B009).
Disclosure
The authors report no conflicts of interest in this work.
References
Fan YH, Xiao B, Lv SG, Ye MH, Zhu XG, Wu MJ. Lentivirus-mediated knockdown of chondroitin polymerizing factor inhibits glioma cell growth in vitro. Oncol Rep. 2017;38(2):1149–1155. | ||
Xiao B, Fan Y, Ye M, et al. Downregulation of COUP-TFII inhibits glioblastoma growth via targeting MPC1. Oncol Lett. 2018;15(6):9697–9702. | ||
Ji CX, Fan YH, Xu F, et al. MicroRNA-375 inhibits glioma cell proliferation and migration by downregulating RWDD3 in vitro. Oncol Rep. 2018;39(4):1825–1834. | ||
Zhang D, Cao C, Liu L, Wu D. Up-regulation of lncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. J Cancer. 2016;7(5):608–617. | ||
Liu J, Liu L, Wan JX, Song Y. Long noncoding RNA SNHG20 promotes gastric cancer progression by inhibiting p21 expression and regulating the GSK-3β/β-catenin signaling pathway. Oncotarget. 2017;8(46):80700–80708. | ||
Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840(3):1063–1071. | ||
Fan Y-H, Fang H, Ji C-Xing, Xie H, Xiao B, Zhu X-G. Long noncoding RNA CCAT2 can predict metastasis and poor prognosis: a meta-analysis. Clin Chim Acta. 2017;466:120–126. | ||
Dai W, Tian C, Jin S. Effect of lncRNA ANRIL silencing on anoikis and cell cycle in human glioma via microRNA-203a. Onco Targets Ther. 2018;11:5103–5109. | ||
Zhou Y, Wang DL, Pang Q. Long noncoding RNA SPRY4-IT1 is a prognostic factor for poor overall survival and has an oncogenic role in glioma. Eur Rev Med Pharmacol Sci. 2016;20(14):3035–3039. | ||
Zheng Y, Gao Y, Li X, et al. Long non-coding RNA NAP1L6 promotes tumor progression and predicts poor prognosis in prostate cancer by targeting Inhibin-β A. Onco Targets Ther. 2018;11:4965–4977. | ||
Liu Q, Liu H, Cheng H, Li Y, Li X, Zhu C. Downregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cells. Onco Targets Ther. 2017;10:2461–2471. | ||
Ma Z, Huang H, Xu Y, et al. Current advances of long non-coding RNA highly upregulated in liver cancer in human tumors. Onco Targets Ther. 2017;10:4711–4717. | ||
Fan Y, Yan T, Chai Y, Jiang Y, Zhu X. Long noncoding RNA HOTTIP as an independent prognostic marker in cancer. Clin Chim Acta. 2018;482:224–230. | ||
Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Guan H, Mei Y, Mi Y, et al. Downregulation of lncRNA ANRIL suppresses growth and metastasis in human osteosarcoma cells. Onco Targets Ther. 2018;11:4893–4899. | ||
Liu H, Pan Y, Han X, Liu J, Li R. MicroRNA-216a promotes the metastasis and epithelial-mesenchymal transition of ovarian cancer by suppressing the PTEN/Akt pathway. Onco Targets Ther. 2017;10:2701–2709. | ||
Fan YH, Ye MH, Wu L, et al. Overexpression of miR-98 inhibits cell invasion in glioma cell lines via downregulation of IKKε. Eur Rev Med Pharmacol Sci. 2015;19(19):3593–3604. | ||
Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–165. | ||
Guan YX, Zhang MZ, Chen XZ, Zhang Q, Liu SZ, Zhang YL. lnc RNA SNHG20 participated in proliferation, invasion, and migration of breast cancer cells via miR-495. J Cell Biochem. 2018;119(10):7971–7981. | ||
He S, Zhao Y, Wang X, et al. Up-regulation of long non-coding RNA SNHG20 promotes ovarian cancer progression via Wnt/β-catenin signaling. Biosci Rep. 2018;38(1):BSR20170681. | ||
Wang W, Luo P, Guo W, et al. lncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem Biophys Res Commun. 2018;503(3):1927–1933. | ||
Li C, Zhou L, He J, Fang X-Q, Zhu S-W, Xiong M-M. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16(1):655. | ||
Munari E, Chaux A, Maldonado L, et al. Cyclin A1 expression predicts progression in pT1 urothelial carcinoma of bladder: a tissue microarray study of 149 patients treated by transurethral resection. Histopathology. 2015;66(2):262–269. | ||
Rodriguez R, Meuth M. Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress. Mol Biol Cell. 2006;17(1):402–412. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





