Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA SLC7A11-AS1 Contributes to Lung Cancer Progression Through Facilitating TRAIP Expression by Inhibiting miR-4775
Authors Liu Y, Fan X, Zhao Z, Shan X
Received 9 March 2020
Accepted for publication 21 May 2020
Published 30 June 2020 Volume 2020:13 Pages 6295—6302
DOI https://doi.org/10.2147/OTT.S253082
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Tohru Yamada
Yongmin Liu,1,* Xinglong Fan,2,* Zheng Zhao,3 Xiu Shan4
1Department of Oncology, General Hospital of Heilongjiang Province Land Reclamation Bureau, Harbin 150088, People’s Republic of China; 2Thoracic Department, Qilu Hospital of Shandong University, Qingdao 266035, People’s Republic of China; 3Third Department of Medical Oncology, Shaanxi Provincial Caner Hospital, Xi’an 710061, People’s Republic of China; 4Department of Oncology, The First Affiliated Hospital of Dalian Medical University, Dalian 116044, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiu Shan
Department of Oncology, The First Affiliated Hospital of Dalian Medical University, Dalian 116044, People’s Republic of China
Email [email protected]
Zheng Zhao
Third Department of Medical Oncology, Shaanxi Provincial Caner Hospital, Xi’an 710061, People’s Republic of China
Email [email protected]
Purpose: Long non-coding RNAs (lncRNAs) are important regulators of lung cancer. This article introduced a novel lncRNA, SLC7A11-AS1, whose effects on lung cancer development have been explored.
Methods: Lung cancer tissues and normal tissues of 47 patients were collected. Bronchial epithelial cell line (BEAS-2B) and lung cancer cell lines (H520, H596, A549 and H1299) were cultured. H1299 and A549 cells were transfected with siSLC7A11-AS1 or siNC. The proliferation, migration and invasion of H1299 and A549 cells were detected by CCK-8 assay and Transwell experiment. Caspase-3 activity in H1299 and A549 cells was researched using caspase-3 activity detection kit. Dual-luciferase reporter gene assay and RNA pull-down assay were performed to explore the relationship between SLC7A11-AS1 and miR-4775. SLC7A11-AS1, miR-4775 and TRAIP mRNA expressions in tissues/cells were detected by qRT-PCR.
Results: The up-regulated SLC7A11-AS1 in lung cancer patients was associated with metastasis and advanced tumor stage (P < 0.05). SLC7A11-AS1 was significantly up-regulated in lung cancer cells (P < 0.05). Silencing of SLC7A11-AS1 prominently inhibited H1299 and A549 cells proliferation, migration and invasion in vitro (P < 0.05). SLC7A11-AS1 acted as a sponge to inhibit miR-4775 expression in H1299 and A549 cells. Meanwhile, TRAIP expression in H1299 and A549 cells was directly and negatively regulated by miR-4775. Inhibition of miR-4775 or overexpression of TRAIP in H1299 and A549 cells remarkably reversed the reduced proliferation, migration and invasion induced by SLC7A11-AS1 silencing (P < 0.05).
Conclusion: SLC7A11-AS1 promoted lung cancer development by enhancing TRAIP expression via suppressing miR-4775.
Keywords: lung cancer, progression, SLC7A11-AS1, miR-4775, TRAIP
Introduction
Lung cancer is one of the major causes of cancer-related deaths with a 5-year survival rate of only 16.1%.1 Small-cell lung cancer and non-small-cell lung cancer are the two subtypes of lung cancer. Among all lung cancer patients, the former accounts for about 15% and the latter accounts for approximately 85%. Non-small-cell lung cancer can be further subdivided into several types, such as squamous cell carcinoma, adenocarcinoma and large cell carcinoma.2 Despite differences among lung cancer subtypes, the delayed diagnosis and limited treatment options are the main reasons for the low survival rate. Given the complicated process of lung cancer development, it is urgently needed to explore novel and specific markers for the diagnosis and target treatment of lung cancer to improve the clinical outcomes.
Long non-coding RNAs (lncRNAs) are a family of RNA molecules with more than 200 nucleotides in length, which have limited protein-coding capacity.3 LncRNAs have been shown to serve functions in multiple human malignant tumors, including lung cancer. Currently, a large amount of lncRNAs have been confirmed to play a vital regulatory role in tumorigenesis of lung cancer.4 For example, a report revealed that LINC00152 was aberrantly up-regulated in lung cancer and high expression of LINC00152 indicated poor prognosis.5 Knockdown of LINC00152 resulted in the weakened proliferation of lung cancer cells.5 On the contrary, lncRNA DGCR5 was reported to be down-regulated in lung cancer and associated with poor prognosis.6 DGCR5 inhibits lung cancer cell proliferation, migration and invasion.6 Several tumor-related lncRNAs have been identified in lung cancer. Thus, discovery of important lncRNAs will provide effective options for the diagnosis and targeted treatment of lung cancer. SLC7A11-AS1 has been rarely reported in human tumors currently. Only two studies identified its function in gastric cancer and pancreatic cancer.7,8
microRNAs (miRNAs) have been identified as crucial regulators in tumorigenesis.9 MiR-4775 has been reported to be participated in the regulation of some human disease, but its function in lung cancer has not yet been reported.10,11 TRAIP was found to promote tumorigenesis by participating in epithelial–mesenchymal transition.12,13 High TRAIP expression has been discovered to be related to metastasis and poor prognosis.12,13 In this article, we have studied the underlying molecular mechanism of SLC7A11-AS1 affecting lung cancer development, which might provide an effective theoretical basis for the application of SLC7A11-AS1 as a potential diagnostic biomarker and therapeutic target.
Methods
Clinical Tissue Samples
Lung cancer tissues and matched normal adjacent tissues of 47 patients were collected. All patients were diagnosed with lung cancer by pathological examination and did not receive radiation or chemotherapy before surgery. Among the 47 lung cancer patients, 26 cases were in stage I/II and 21 cases were in stage III/IV. Meanwhile, 22 patients were with metastasis and the other 25 patients were without metastasis. In this study, all participants have signed written informed consent. This research has been approved by the ethics committee of the First Affiliated Hospital of Dalian Medical University in line with the Helsinki Declaration.
Cell Culture
Bronchial epithelial cell line (BEAS-2B) and lung cancer cell lines (H520, H596, A549 and H1299) were provided by Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco′s Modified Eagle′s Medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C, 5% CO2 in a sterile incubator.
Transfection
Specific SLC7A11-AS1 interfering oligonucleotides (si-SLC7A11-AS1-1 and si-SLC7A11-AS1-2) and negative control oligonucleotides (si-NC) were synthesized by Ribo (Guangzhou, China) and transfected into H1299 and A549 cells. MiR-4775 mimics, miR-4775 inhibitors and negative controls were obtained from GenePharma (Shanghai, China). Transfection was performed using Lipofectamine 2000 (Invitrogen, USA) in strict accordance with the instructions. After 48 h of transfection, cells were collected for subsequent researches.
CCK-8 Assay
The transfected H1299 and A549 cells were dispersed into single-cell suspensions. Each single-cell suspension was added into 96-well plates. Cells in each plate were cultured for 24 h, 48 h and 72 h, respectively, at 37°C, 5% CO2. At each time point, the plates were removed and CCK-8 reagent with a volume of 10 μL was added to incubate cells for 4 h at 37°C. The optical density (OP) value of cells was measured by a microplate reader (Biotek, USA).
Transwell Experiment
After transfected, cell migration and invasion ability were researched by Transwell experiment. In short, the Transwell inserts were inserted into 24-well plates. Matrigel was spread onto the upper chamber for invasion assay. The transfected H1299 and A549 cells dispersed in serum-free DMEM were added into the upper chamber. In the lower chamber, DMEM containing 10% FBS was added with a volume of 500 μL. These 24-well plates were incubated in the incubator at 37°C, 5% CO2 for 24 h. Cells on the upper surface were removed by cotton swab, and those on the lower surface was fixed with paraformaldehyde (4%) and stained with crystal violet (0.1%). The number of invasive cells was counted under an inverted microscope. According to the above operation, the number of migration cells was also counted. It should be noted that Matrigel was not required in the determination of migration capacity detection.
Detection of Caspase-3 Activity
The transfected H1299 and A549 cells were seeded in 6-well plates. Cells in all plates were cultured for 24 h at 37°C, 5% CO2. Then, cells were collected and the activity of caspase-3 was investigated using caspase-3 activity assay buffer solution. The caspase-3 activity detection kit was provided by Beyotime Institute of Biotechnology (Shanghai, China).
Dual-Luciferase Reporter Gene Assay
The wild type (WT)-SLC7A11-AS1, mutant type (MUT)-SLC7A11-AS1, WT-TRAIP and MUT-TRAIP containing the binding site for miR-4775 were designed and inserted into pGL3 luciferase reporter (Promega). For luciferase reporter assay, H1299 and A549 cells transfected by miR-4775 mimics and negative control were further subjected to co-transfection with the above luciferase reporter vectors. After 48 h of co-transfection, dual-luciferase reporter assay kit (Promega, USA) was used to detect the luciferase activity of cells and the relative luciferase activity was calculated.
qRT-PCR
The expression level of SLC7A11-AS1, miR-4775 and TRAIP mRNA in tissues and cells were determined by qRT-PCR. Briefly, total RNA was extracted from tissues and cells using TRIzol Reagent. Equal amount of each RNA sample was subjected to reverse transcription reaction to synthesize cDNA templates by using PrimeScript™ RT reagent kit (Takara, Japan) in strict accordance with the manual. The cDNA templates were then quantified with SYBR Green reagent (Takara, China) by using the ABI 7500 fast real-time PCR System (Applied Biosystems, USA). The amplification reaction conditions were as follows: denaturation at 94°C for 5 min, 40 cycles of 94°C for 30 s, 58°C for 45 s and 72°C for 30 s, followed by extension at 72°C for 10 min. The relative expression of SLC7A11-AS1, miR-4775 and TRAIP mRNA was calculated with the 2−ΔΔCt method with U6 as the internal control for SLC7A11-AS1, miR-4775 and GAPDH as the internal control for TRAIP mRNA.
Statistical Analysis
Data were expressed in the form of mean and standard deviation and analyzed by SPSS 21.0 software. Student’s t test was used for the comparison between two groups, while one-way analysis of variance was applied to perform comparison at least three groups. P < 0.05 was considered as statistically significant difference. All experiments were repeated at least three times.
Results
The Expression of SLC7A11-AS1 Was Up-Regulated in Lung Cancer Patients and Cells
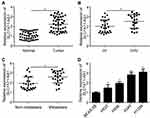
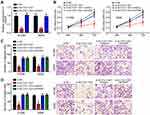
SLC7A11-AS1 relative expression in tumor tissues and normal tissues of 47 lung cancer patients was detected by qRT-PCR. The results showed that relative expression of SLC7A11-AS1 was up-regulated in tumor tissues (Figure 1A). Besides, SLC7A11-AS1 expression in tumor tissues of patients with stage III/IV was higher than those in stage I/II (P < 0.05) (Figure 1B). Meanwhile, tumor tissues with metastasis had much higher SLC7A11-AS1 expression (Figure 1C). SLC7A11-AS1 expression in lung cancer cell lines was subsequently researched, and data showed that SLC7A11-AS1 expression was up-regulated in four lung cancer cell lines (H520, H596, A549 and H1299) compared to that in BEAS-2B cell line (P < 0.05) (Figure 1D).
SLC7A11-AS1 Silencing Inhibited Proliferation, Migration and Invasion of Lung Cancer Cells in vitro
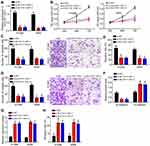
Specific SLC7A11-AS1 interfering oligonucleotides (si-SLC7A11-AS1-1 and si-SLC7A11-AS1-2) and negative control oligonucleotides (si-NC) were transfected into H1299 and A549 cells. By transfection, the relative SLC7A11-AS1 expression in H1299 and A549 cells of si-SLC7A11-AS1-1 group and si-SLC7A11-AS1-2 group was both markedly decreased, indicating that SLC7A11-AS1 expression was effectively silenced (Figure 2A). Then, the effect of SLC7A11-AS1 silencing on cell phenotype was investigated. As shown by CCK8 and Transwell assays, SLC7A11-AS1 knockdown significantly inhibited proliferation, migration and invasion (Figure 2B–D). Besides, the wound healing assay further validated that SLC7A11-AS1 silencing suppressed cell migration (Figure 2E). Moreover, SLC7A11-AS1 silencing promoted E-cadherin expression while inhibiting the level of N-cadherin (Figure 2F), suggesting that SLC7A11-AS1 knockdown prevents epithelial-to-mesenchymal transition (EMT) progression. In addition, the activity of caspase-3 was explored. Compared with si-NC group, H1299 and A549 cells of si-SLC7A11-AS1-1 group and si-SLC7A11-AS1-2 group showed much higher relative caspase-3 activity (P < 0.05) (Figure 2G), indicated that SLC7A11-AS1 silencing induced apoptosis. Flow cytometry result also demonstrated that SLC7A11-AS1 silencing promoted apoptosis (Figure 2H).
SLC7A11-AS1 Directly Bound to miR-4775 and Regulated Its Expression Negatively
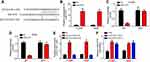
Through bioinformatics analysis using miRDB (http://mirdb.org/), we found that SLC7A11-AS1 may be the sponge for miR-4775 which scored the highest. To validate it, the WT-SLC7A11-AS1 and MUT-SLC7A11-AS1 sequences containing the miR-4775 binding site were synthesized to generate luciferase reporter vectors (Figure 3A). The relationship between miR-4775 and SLC7A11-AS1 was verified subsequently by luciferase reporter assay. H1299 and A549 cells were transfected by miR-4775 mimics (miR-4775 group) and negative control (NC group). Obviously, miR-4775 mimic transfection effectively promoted miR-4775 expression (Figure 3B). Luciferase reporter assay showed that the relative luciferase activity of WT-SLC7A11-AS1 reporter in miR-4775 group was prominently inhibited (P < 0.05), but that of MUT-SLC7A11-AS1 reporter in miR-4775 group was not obviously changed (Figure 3C and D). Besides, data from RNA pulldown assay showed that WT-bio-miR-4775 group significantly enriched SLC7A11-AS1 (Figure 3E), suggesting SLC7A11-AS1 directly interacted with miR-4775. Furthermore, SLC7A11-AS1 knockdown promoted the levels of miR-4775 in H1299 and A549 cells (Figure 3F).
MiR-4775 Could Target TRAIP and Regulated Its Expression Negatively
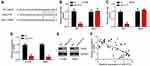
Then, bioinformatics analyses were performed to identify the target of miR4775 by using miRDB and TargetScan tools. Results showed that TRAIP ranked top. Then, the WT-TRAIP and MUT-TRAIP sequences containing the binding site for miR-4775 were synthesized to generate luciferase reporter vectors (Figure 4A). Dual-luciferase reporter assay exhibited that the relative luciferase activity of WT-RAIP reporter was markedly weakened by miR-4775 mimics (P < 0.05), but that of MUT-TRAIP reporter was not obviously affected by miR-4775 mimics (Figure 4B and C). Furthermore, miR-4775 mimics remarkably inhibited TRAIP expression (Figure 4D and E). Obviously, in tumor tissues of lung cancer patients, TRAIP expression was negatively correlated with the expression of miR-4775 (P < 0.05) (Figure 4F).
SLC7A11-AS1 Could Promote Lung Cancer Development by Enhancing TRAIP Expression via Suppressing miR-4775
Interestingly, we found that SLC7A11-AS1 knockdown suppressed TRAIP expression and it is reversed by miR-4775 inhibitors (Figure 5A). To validate the regulatory roles of SLC7A11-AS1/miR-4775/TRAIP axis in lung cancer, rescue assays were carried out. Through CCK8 and Transwell assays, we found that miR-4775 inhibition or TRAIP overexpression rescued the proliferation, migration and invasion of lung cancer cells with SLC7A11-AS1 knockdown (Figure 5B–D). Therefore, SLC7A11-AS1 could promote lung cancer development by enhancing TRAIP expression through suppressing miR-4775.
Discussion
Great progress has been achieved in the diagnosis and treatment of lung cancer in the past few decades, but its low 5-year survival rate remains a difficult problem to be solved worldwide.1 LncRNAs have been proved to be involved in the regulation of tumorigenesis.14,15 In this article, we introduced SLC7A11-AS1 as a potential prognostic marker and therapeutic target for lung cancer. Through binding to miR-4775, SLC7A11-AS1 could promote the expression of TRAIP to enhance lung cancer cell proliferation, migration and invasion.
In previous study, SLC7A11-AS1 was noticed to be down-regulated in gastric cancer and correlated with poor outcomes.7 SLC7A11-AS1 was suggested to be a useful biomarker for gastric cancer diagnosis.7 On the contrary, SLC7A11-AS1 was proved to be abnormally up-regulated in patients with pancreatic cancer.8 However, the role of SLC7A11-AS1 in lung cancer remains unknown. For the first time, our study illustrated the up-regulated SLC7A11-AS1 in lung cancer was associated with tumor progression. SLC7A11-AS1 knockdown suppressed the proliferation, migration and invasion of lung cancer cells.
LncRNAs have been reported to be ceRNAs for miRNAs.11 And, miRNAs have been considered as powerful regulators of human tumors by affecting multiple biological processes such as proliferation, migration and invasion. Through bioinformatics analysis, we found that SLC7A11-AS1 was the sponge for miR-4775. We demonstrated the interaction between miR-4775 and SLC7A11-AS1. MiR-4775 was previously found to be a tumor suppressor in glioma and its expression was obviously declined in glioma tissues.11 MiR-4775 overexpression significantly reduced glioma cell proliferation and migration in vitro.11 Furthermore, Zhao et al illustrated that miR-4775 promotes colorectal cancer growth, invasiveness and epithelial–mesenchymal transition.10 Nevertheless, how miR-4775 regulates lung cancer is elusive. Results from our study revealed that miR-4775 expression in lung cancer was negatively regulated by SLC7A11-AS1. And miR-4775 could serve as an important tumor suppressor in lung cancer.
Through bioinformatics analysis, we further identified and proved that TRAIP was directly targeted by miR-4775. And we found that TRAIP expression was regulated by SLC7A11-AS1/miR-4775 axis. The function of TRAIP in cancer is rarely researched. Only a study showed that TRAIP could drive the epithelial–mesenchymal transition of liver cancer and TRAIP overexpression was closely correlated with metastasis and poor prognosis.16 However, the function of TRAIP in lung cancer is largely unclear. In this research, we found that TRAIP expression was directly inhibited by miR-4775 and overexpression of TRAIP remarkably reversed the reduced proliferation, migration and invasion induced by SLC7A11-AS1 silencing. Thus, TRAIP was an oncogene in lung cancer. However, whether SLC7A11-AS1/miR-4775/TRAIP axis regulates the activation of some classical pathways, such as Wnt signaling, requires more investigation.
In summary, this research demonstrated that SLC7A11-AS1 was prominently up-regulated in lung cancer. SLC7A11-AS1 could promote lung cancer development by enhancing TRAIP expression via suppressing miR-4775. This study is the first time to discover the function of SLC7A11-AS1 in lung cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Guo Y, Hu Y, Hu M, He J, Li B. Long non-coding RNA ZEB2-AS1 promotes proliferation and inhibits apoptosis in human lung cancer cells. Oncol Lett. 2018;15(4)5220–5226.
2. Liu BY, Ye BQ, Yang LL, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499–508. doi:10.1038/ni.3712
3. Yang Q, Jia L, Li X, Guo R, Li W. Long noncoding RNAs: new players in the osteogenic differentiation of bone marrow- and adipose-derived mesenchymal stem cells. Stem Cell Rev Rep. 2018;14(7414):1–12. doi:10.1007/s12015-018-9801-5
4. Duan C, Chunfang Z, Zhenzi P. Functions and mechanisms of long noncoding RNAs in lung cancer. Chin J Clin Oncol. 2016;9:4411–4424.
5. Feng S, Zhang J, Su W, Bai S, Chen G. Overexpression of LINC00152 correlates with poor patient survival and knockdown impairs cell proliferation in lung cancer. Sci Rep. 2017;7(1):2982. doi:10.1038/s41598-017-03043-x
6. Chen EG, Zhang JS, Xu S, Zhu XJ, Hu HH. Long non-coding RNA DGCR5 is involved in the regulation of proliferation, migration and invasion of lung cancer by targeting miR-1180. Am J Cancer Res. 2017;7(7):1463–1475.
7. Luo Y, Wang C, Yong P, et al. Decreased expression of the long non-coding RNA SLC7A11-AS1 predicts poor prognosis and promotes tumor growth in gastric cancer. Oncotarget. 2017;8(68):112530–112549. doi:10.18632/oncotarget.22486
8. Yang Q, Li K, Huang X, et al. lncRNA SLC7A11-AS1 promotes chemoresistance by blocking SCF (beta-TRCP)-mediated degradation of NRF2 in pancreatic cancer. Mol Ther Nucleic Acids. 2020;19:974–985. doi:10.1016/j.omtn.2019.11.035
9. Zhu B, Shaoqing JU, Chu H, Shen X, Zhang Y. The potential function of microRNAs as biomarkers and therapeutic targets in multiple myeloma (Review). Oncol Lett. 2018;15(5):6094–6106. doi:10.3892/ol.2018.8157
10. Zhao S, Sun H, Jiang W, et al. miR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFbeta-mediated epithelial to mesenchymal transition. Mol Cancer. 2017;16(1):12.
11. Zhu Z, Dai J, Liao Y, Ma J, Zhou W. Knockdown of long noncoding RNA LINC00152 suppresses cellular proliferation and invasion in glioma cells by regulating miR-4775. Oncol Res. 2018;26(6):857–867. doi:10.3727/096504017X15016337254597
12. Graham TR, Zhau HE, Oderomarah VA, et al. Insulin-like growth factor-i–dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008;27(1):2479–2488.
13. Sanchez-Tillo E, de Barrios O, Siles L, et al. ZEB1 promotes invasiveness of colorectal carcinoma cells through the opposing regulation of uPA and PAI-1. Clin Cancer Res. 2013;19(5):1071–1082. doi:10.1158/1078-0432.CCR-12-2675
14. Chen Y, Xie H, Gao Q, et al. Colon cancer associated transcripts in human cancers. Biomed Pharmacother. 2017;94:531–540. doi:10.1016/j.biopha.2017.07.073
15. Pentenero M, Bowers L, Jayasinghe R, Cheong SC, Alevizos I. World workshop on oral medicine VII: functional pathways involving differentially expressed lncRNAs in oral squamous cell carcinoma. Oral Dis. 2019;25(S1):79–87. doi:10.1111/odi.13051
16. Guo Z, Zeng Y, Chen Y, et al. TRAIP promotes malignant behaviors and correlates with poor prognosis in liver cancer. Biomed Pharmacother. 2020;124:109857. doi:10.1016/j.biopha.2020.109857
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.