Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 18
LncRNA RP11-521C20.3 Inhibits Cigarette Smoke Extract-Induced Apoptosis in A549 Cells by Targeting BMF Signaling
Authors Zhong Y, Li C, Xiang Y, Zhou J, Zhang J
Received 22 November 2022
Accepted for publication 7 March 2023
Published 21 April 2023 Volume 2023:18 Pages 669—682
DOI https://doi.org/10.2147/COPD.S395568
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Yong Zhong, Chuntao Li, Yaling Xiang, Jinbiao Zhou, Jianqing Zhang
Department of Respiratory and Critical Care Medicine, the First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan, People’s Republic of China
Correspondence: Jianqing Zhang, Department of Respiratory Critical Care Medicine, the First Affiliated Hospital of Kunming Medical University, Kunming, Yunnan, 650032, People’s Republic of China, Tel +86 18988272502, Email [email protected]
Objective: LncRNAs are closely correlated with chronic obstructive pulmonary disease (COPD). We investigated the molecular mechanism of lncRNA RP11-521C20.3, which targets the action of the Bcl-2 modifying factor (BMF) signaling pathway in the apoptosis of cigarette smoke extract (CSE)-treated A549 cells.
Methods: Lung tissues derived from cigarette smoke exposed rats (COPD group) and controls were examined using TUNEL assay for apoptotic cells and using immunohistochemistry for BMF expression levels. Overexpression and knockdown of BMF by lentiviral vector transfection were used to explore the role of BMF on the apoptosis of CSE-treated A549 cells. Overexpression and knockdown of RP11-521C20.3 were used to assess the effect of RP11-521C20.3 on the expression levels of BMF and apoptosis in CSE-treated A549 cells. Cell proliferation, mitochondrial morphology, and apoptosis were assessed in A549 cells. Real-time quantitative polymerase chain reactions and Western blotting detected the expression of apoptosis-related molecules.
Results: The number of apoptotic cells and the level of BMF protein were significantly increased in lung tissues of the COPD group compared to the control group. Overexpression of BMF or knockdown of RP11-521C20.3 in CSE-treated A549 cells increased apoptosis, inhibited cell proliferation, and exacerbated mitochondrial damage. There were also increased protein levels of p53, cleaved caspase-3, and cleaved caspase-7, and decreased protein levels of Bcl-2 and survivin. Knockdown of BMF or overexpression of RP11-521C20.3 in CSE-treated A549 cells attenuated apoptosis, promoted cell proliferation, and alleviated mitochondrial damage. Observed effects also included decreased protein levels of p53, cleaved caspase-3, and cleaved caspase-7, and increased protein levels of Bcl-2 and survivin. In CSE-treated A549 cells, overexpression of RP11-521C20.3 suppressed the expression of BMF mRNA and protein.
Conclusion: In CSE-treated A549 cells, BMF promoted apoptosis and RP11-521C20.3 might target the BMF signaling axis to protect CSE-treated A549 cells from apoptosis.
Keywords: chronic obstructive pulmonary disease, lncRNA RP11-521C20.3, BMF, apoptosis
Introduction
Chronic obstructive pulmonary disease (COPD) is a preventable and treatable respiratory disease but is incurable.1 Cigarette smoke (CS) remains the most common and important environmental risk factor for COPD.2,3 Smoking-mediated oxidative stress or inflammatory responses triggered by smoking can cause lung tissue damage.4 It has been proposed that COPD develops and progresses due to increased apoptosis of alveolar epithelial cells and pulmonary vascular endothelial cells, resulting in lung damage.5,6 However, the molecular mechanism of CS-induced apoptosis in COPD has not been fully elucidated.
Apoptosis removes abnormal, damaged, and unwanted cells and plays an important role in tissue homeostasis and immunity.7,8 Apoptosis occurs through both extrinsic and intrinsic pathways.8,9 Apoptosis is regulated by members of the Bcl-2 family, the inhibitor of apoptosis proteins family and the caspase family. Bcl-2, an anti-apoptotic protein, inhibits cell death by suppressing the function of pro-apoptotic proteins in the Bcl-2 family.8 Survivin is a member of the mammalian inhibitor of apoptosis proteins family, which inhibits caspase-dependent and non-caspase-independent apoptosis.10 Caspase-3 and caspase-7 are members of a conserved family of cysteine proteases (caspases).11 Activation of caspase-3 and caspase-7 promotes apoptosis.11 The cytoplasmic form of the tumor suppressor protein p53 translocates to the mitochondria and interacts with members of the Bcl-2 family to induce apoptosis.12,13
Bcl-2 modifying factor (BMF) is a member of the Bcl-2 family of pro-apoptotic BH3-only proteins.14 Under normal conditions, BMF is sequestered in the cellular cytoskeleton through interactions with kinesin light chain 2 via the myosin V motor complex.15 When cells are subjected to certain stressful stimuli, BMF is released from the cytoskeleton and localizes to the mitochondria to initiate pro-apoptotic activity.15–18 Levels of BMF mRNA are elevated in the peripheral blood of COPD patients compared to healthy individuals who smoke.19 Current studies on BMF regulation of apoptosis are primarily focused on cancer and autoimmune diseases.20–22 Studies on the functional apoptotic mechanism of BMF in COPD have not been reported.
Tobacco smoke exposure can alter the expression and function of long non-coding RNAs(lncRNAs), which may be involved in the onset and progression of COPD.23,24 Even heat-not-burn tobacco (IQOS) smoke seems to affect the proliferation of human gingival fibroblasts and human keratinocytes.25 LncRNA SNHG5 increases PTEN expression by sponging miR-132 to attenuate the influence of cigarette smoke extract (CSE) on cellular proliferation and apoptosis, and tissue inflammation.26 The lncRNA MEG3 targets different microRNAs to regulate the expression of protein-coding genes, ultimately regulating the effects of CSE on cellular proliferation and apoptosis, and tissue inflammation.27–29
The expression of the lncRNA RP11-521C20.3 (BMF antisense RNA 1, BMF-AS1) is reduced in the peripheral blood of COPD patients and the expression of its associated BMF mRNA is elevated.19 Many natural antisense transcripts have been described as lncRNAs,30 which mediate the regulation of sense mRNA through various mechanisms such as transcriptional interference, epigenetic modifications, processing of RNA and selective splicing.31 Therefore, RP11-521C20.3 may regulate the expression of BMF and thereby play an important apoptotic function in COPD.
We investigated the biological function of RP11-521C20.3 and its regulatory effect on BMF. We also investigated the pathological effects of BMF, its biological function, and its regulatory role on downstream molecules in apoptosis.
Materials and Methods
Sample Preparation
The Experimental Zoology Center of Kunming Medical University provided 30 healthy male and female Sprague-Dawley rats weighing 130–160 g. CS exposed rats were constructed as described previously.32,33 They were randomly divided into control and smoking groups, reared on standard chow, and watered ad libitum. Rats were housed in a controlled experimental environment (temperature: 22 ± 2 °C; light hours: 6:00–18:00). The rats in the smoking group were placed in a PAB-S200 Animal Passive Smoking Exposure System (BioLab Technology Co. Ltd., Beijing, China), which consists of five parts: passive smoking smoke poisoning box, cigarette smoke generator, gas sensor, signal conditioner, real-time data recording and analysis software. The rats exposed to CS once per day: 5 sets of 3 cigarettes/15 min with a 3-min pause between each set of 3 cigarettes (Per cigarette contains 1.0 mg of nicotine, 11 mg of tar), 6 d/wk for 4 mo. The control group was exposed to room air. Rats were executed after the last CS exposure.
Lung Histology
Rat lung tissues were fixed with 4% paraformaldehyde, embedded in paraffin, cut into 3-4-μm-thick sections, and stained with hematoxylin and eosin (HE).
Immunohistochemistry
Fixed rat lung tissue slides were dewaxed, hydrated, and washed with PBS. The slides were incubated with 3% hydrogen peroxide for 10–20 min in the dark and washed with PBST buffer. The slides were incubated with 10% goat serum for 20–60 min, washed, and incubated with a BMF primary antibody (Catalog # ab9655, Abcam, Shanghai, China) at a dilution of 1:100 at 4 °C overnight. Subsequently, 50 μL of enhanced enzyme-labeled goat anti-mouse/rabbit IgG polymer was added, and the slides were incubated for 20 min at room temperature (RT). The slides were washed in tap water, and the color was developed with 50 μL of DAB working solution. The slides were re-stained with hematoxylin and dehydrated, transparent, and sealed. The slides were observed under 400X microscopy and photographed. Images of three slides were taken from each of the experimental and control groups, and the images were analyzed using Image-Pro Plus image analysis software to measure the integral optical density (IOD) values of positive BMF expression. Five visual fields were randomly selected for each slide, and the mean optical density (MOD) values of the five visual fields were used as the relative expression level of BMF in the samples.
TUNEL and DAPI Staining
Rat lung tissues were paraffin-embedded and sectioned for Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) staining (No. KGA7063, KeyGEN BioTECH, Nanjing, China). The slides were treated with a proteinase K working solution for 10 min at RT and washed with PBS. A TdT enzyme reaction solution (50 μL) was added dropwise to the slides and incubated at 37 °C for 60 min in the dark, then the slides were washed with PBS. A streptavidin-TRITC labeling solution (50 μL) was added dropwise to the slides, and the slides were incubated at 37 °C for 30 min in the dark, then washed with PBS. The slides were incubated with a DAPI solution (50 μL) for 10 min, then washed with PBS. The slides were observed under fluorescent microscopy and photographed. Three slides were selected for each group, and four fields of view were randomly selected for each slide. The positive points of DAPI and Tunel were counted separately using Image J software. The positive points of DAPI were taken as the total number of cells, the positive points of TUNEL were taken as the number of apoptotic cells, and the apoptosis rate (%) = TUNEL/DAPI×100%. The apoptosis rate of this sample was the average of the apoptosis rates of the four fields of vision.
Cigarette Smoke Extract Preparation
CSE was prepared as previously described.34 Two commercially available unfiltered cigarette brands (Hongyun Honghe Tobacco, Yunnan, China) were burned, and the smoke was drawn through 25 mL of serum-free Ham’s F-12K medium (Procell, Wuhan, China) using a vacuum pump. The medium was filtered through a 0.22-μm filter, and the pH was adjusted to 7.2–7.4. The final preparation was considered a 100% CSE solution. CSE was stored at −80°C until needed. CSE was diluted with complete Ham’s F-12K medium to make working concentrations of 0.5%, 1%, 2.5%, 5%, 10%, 20%, and 40%.
Lentivirus Production and Transfection
Recombinant lentiviral vectors overexpressing RP11-521C20.3, BMF and the corresponding negative controls were designed and constructed (Obio Technology, Shanghai, China). The recombinant lentiviral vectors expressing short hairpin RNA (shRNA) targeting RP11-521.C20.3, BMF and the corresponding negative controls were also purchased from Obio Technology. A549 cells were inoculated in six-well microtiter plates at a density of 1×105 cells/well. When the cells reached 30%-40% confluency, they were transfected with recombinant lentiviruses at a multiplicity of infection of 5. After a 24 h transfection period, the supernatant was replaced with fresh medium. After 72 h, successfully transfected cells were selected using 2 μg/mL puromycin. Inverted fluorescence microscopy (Mshot, Guangzhou, China) confirmed the transfection efficiency, and RT-qPCR was used to determine the expression level of the target genes. The primer sequences used are shown in Table 1.
 |
Table 1 Primer Sequences Used in the Experiments |
Cell Proliferation Assay
Transfected and control A549 cells were treated with 10% CSE for 24 h. Cells from the different treatment groups were seeded in 96-well plates at a density of 3×103 cells/well. Cell proliferation was determined using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Rockville, MD). At 0, 24, 48, and 72 h after inoculation, the medium was removed from the cells, 10 μL of CCK-8 reagent was added to each well, and the cells were incubated at 37 °C for 2 h. The optical density (OD) at 450 nm was determined using an RT-2100C microplate reader (Rayto Life and Analytical Sciences, Shenzhen, China). The OD value of each experimental group was equal to the measured OD value of the experimental group minus the OD value of the blank group.
Cell Apoptosis Assay
Apoptosis was measured using an Annexin V-kFluor647/PI apoptosis detection kit (KeyGEN Biotech, Nanjing, China) according to the manufacturer’s instructions. Control or untransfected and transfected CSE-treated A549 cells were collected and centrifuged and then washed 3 times with cold PBS. Cells were resuspended and stained with Annexin V and propidium iodide for 20 min in the dark at RT. The number of apoptotic cells (10,000 analyzed cells/per sample) was detected within 1 h using the BD Accuri C6 Plus flow cytometer (BD Bioscience, Shanghai, China). We used Flowjo software (https://www.flowjo.com/solutions/flowjo, ver. 10.6.2) to analyze the data.
RT-qPCR
Total RNA was extracted from cells of different treatment groups using TriQuick Reagent (Solarbio Life Science, Beijing, China), chloroform, isopropanol, and 75% ethanol. Total RNA was reverse-transcribed to cDNA using the Servicebio® RT First Strand cDNA Synthesis Kit (Servicebio Technology, Wuhan, China). qPCR was performed using the 2×SYBR Green qPCR Master Mix (Servicebio Technology, Wuhan, China) in an Applied Biosystem’s StepOnePlus real-time PCR system (Thermo Fisher Scientific, Shanghai, China). GAPDH was used as a standardized control. The sequences of the primers (Generay Biotech, Shanghai, China.) are shown in Table 1.
Transmission Electron Microscopy
Transfected and untransfected A549 cells were treated with 10% CSE for 24 h. Cells were fixed in glutaraldehyde for electron microscopy. Sections were observed with a JEM-1400Plus transmission electron microscope (JEOL, Beijing, China).
Western Blotting Analysis
Transfected and untransfected A549 cells were treated with 10% CSE for 24 h. A549 cells were collected and lysed with Radio Immunoprecipitation Assay Lysis buffer (Solarbio Life Science, Beijing, China) containing PMSF on ice for 30 min. The supernatant was obtained by centrifugation at 4 °C and 12 000 rpm for 20 min to obtain total protein. The protein concentration was measured by a BCA protein assay kit (Solarbio Life Science, Beijing, China). The loading amount of each sample was determined according to the concentration of the protein and the gray value of GAPDH. Equal amounts of protein samples were separated using SDS gel electrophoresis and transferred to PVDF membranes (Millipore Sigma, Beijing, China). The membranes were cut according to the molecular weight of each target protein during the transfer. The membranes were blocked with 5% skim milk for 1 h at RT and incubated with Abcam (Shanghai, China) primary antibodies for BMF (ab9655), Bcl-2 (ab32124), p53 (ab26), survivin (ab76424), cleaved caspase-3 (ab32042), cleaved caspase-7 (ab256469), and GADPH (ab8245) at 4 °C overnight. Membranes were then incubated with secondary antibodies at RT for 2 h. Incubation with the ECL reagent (Millipore, Sigma Beijing, China) revealed protein bands, which were observed using a Chemidoc XRS+ chemiluminescence imaging system (Bio-RAD, Beijing, China). Image J software (https://imagej.nih.gov/ij/download.html) was used to quantify the scanned images.
Statistical Analysis
Data were analyzed using SPSS 20.0 (https://www.ibm.com/support/pages/spss-statistics-20-available-download) and GraphPad Prism 7.0 software (GraphPad, San Francisco, CA). Data are expressed as mean plus and minus standard deviation (SD). Unpaired Student’s t test and one-way ANOVA were used to assess statistical differences between 2 and multiple groups, respectively. P < 0.05 was considered statistically significant.
Results
Increased Apoptosis and Elevated Levels of BMF Protein in Lung Tissue of COPD Rats
HE staining of lung tissues in the control group showed no obvious pathological changes, while alveolar expansion was observed in the lung tissues of the COPD group (Figure 1A). Alveolar expansion included thinning and breaking of alveolar septa in some areas and the fusion of the expanded alveoli into larger cystic cavities (Figure 1A). The number of apoptotic cells was significantly increased in the lungs of the COPD group compared to normal controls (Figure 1B and Figure 1). Immunohistochemical analysis showed that the mean number of BMF-positive cells was significantly higher in the COPD group than that in the control group (Figure 1D and Figure 1). These data suggest that BMF was associated with the apoptosis of alveolar epithelial cells in COPD.
CSE Promoted Apoptosis in A549 Cells
The percentage of apoptotic A549 cells was significantly increased in the 10%, 20%, and 40% CSE-treated groups compared to the control medium group (Figure 2). Among them, 10% CSE significantly induced apoptosis, and all subsequent A549 experiments were treated with 10% CSE for 24 h.
BMF Promoted Apoptosis in CSE-Treated A549 Cells
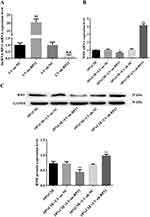
We overexpressed or knocked down BMF using lentiviral transfection in A549 cells (Figure 3A). Next, we investigated cell proliferation, apoptosis, mitochondrial integrity, and mRNA and protein levels of apoptosis-related molecules in transfected and untransfected CSE-treated A549 cells. Overexpression of BMF in CSE-treated A549 cells inhibited proliferation and exacerbated apoptosis compared to untransfected controls (Figure 3B and C). These cells were characterized by disrupted mitochondrial double-membrane structures, decreased number of intra-mitochondrial cristae, severe vacuolization, and the appearance of autophagosomes (Figure 3D). Overexpression of BMF in CSE-treated A549 cells increased the levels of BMF and p53 mRNA and decreased the levels of Bcl-2 and survivin mRNA but had no effect on the expression of caspase-3 and caspase-7 mRNA compared to untransfected controls (Figure 3E). Overexpression of BMF in CSE-treated A549 cells increased the protein levels of BMF, p53, cleaved caspase-3, and cleaved caspase-7, and decreased the protein levels of Bcl-2 and survivin (Figure 3F).
Figure 3 Continued.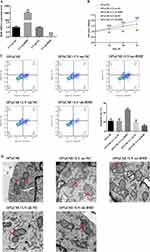
Knockdown of BMF expression in CSE-treated A549 cells promoted proliferation and attenuated apoptosis compared to untransfected controls (Figure 3B and C). These cells were characterized by intact mitochondrial double-membrane structures, uniform distribution of intra-mitochondrial cristae, and slight vacuolization compared to untransfected controls (Figure 3D). Knockdown of BMF expression in CSE-treated A549 cells decreased the levels of BMF and p53 mRNA, elevated the levels of Bcl-2 and survivin mRNA, and had no effect on the levels of caspase-3 and caspase-7 mRNA (Figure 3E). Knockdown of BMF expression in CSE-treated A549 cells decreased the protein levels of BMF, p53, cleaved caspase-3, and cleaved caspase-7, and increased the protein levels of Bcl-2 and survivin (Figure 3F). The above data indicated that BMF promoted CSE-induced apoptosis in CSE-treated A549 cells by increasing the expression of pro-apoptotic mediators (p53, caspase-3, and caspase-7) and inhibiting the expression of anti-apoptotic mediators (Bcl-2 and survivin).
RP11-521C20.3 Inhibited the Expression of BMF in CSE-Treated A549 Cells
We overexpressed or knocked down RP11-521C20.3 using lentiviral transfection in A549 cells (Figure 4A). In CSE-treated A549 cells overexpressing RP11-521C20.3, we observed decreased expression of BMF at both the mRNA and protein levels compared to untransfected controls (Figure 4B and Figure 4). Knockdown of RP11-521C20.3 in CSE-treated A549 cells increased the expression of BMF at the mRNA and protein levels compared to untransfected controls (Figure 4B and Figure 4). This suggests that RP11-521C20.3 negatively regulated the expression of BMF.
RP11-521C20.3 Attenuated Apoptosis in CSE-Treated A549 Cells
Overexpression of RP11-521C20.3 in CSE-treated A549 cells promoted proliferation and reduced apoptosis (Figure 5A and B). These cells were characterized by intact mitochondrial double-membrane structures, uniform distribution of intra-mitochondrial cristae, and slight vacuolization compared to untransfected controls (Figures 5C). Overexpression of RP11-521C20.3 in CSE-treated A549 cells decreased the level of p53 mRNA and increased the levels of Bcl-2 and survivin mRNA but had no effect on the levels of caspase-3 and caspase-7 mRNA compared to untransfected controls (Figure 5D). Overexpression of RP11-521C20.3 in CSE-treated A549 cells decreased the protein levels of p53, cleaved caspase-3, and cleaved caspase-7, and increased the protein levels of Bcl-2 and survivin compared to untransfected controls (Figure 5E).
Figure 5 Continued.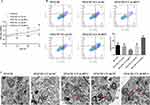
Knockdown of RP11-521C20.3 expression in CSE-treated A549 cells inhibited proliferation and exacerbated apoptosis (Figure 5A and B). These cells were characterized by swollen and disrupted mitochondrial double-membrane structures, a significantly reduced number of intra-mitochondrial cristae, severe vacuolization, and the appearance of autophagosomes compared to untransfected controls (Figure 5C). Knockdown of RP11-521C20.3 expression in CSE-treated A549 cells increased the level of p53 mRNA and decreased the levels of Bcl-2 and survivin mRNA but had no effect on the levels of caspase-3 and caspase-7 mRNA compared to untransfected controls (Figure 5D). Knockdown of RP11-521C20.3 expression in CSE-treated A549 cells elevated the protein levels of p53, cleaved caspase-3, and cleaved caspase-7, and decreased the protein levels of Bcl-2 and survivin compared to untransfected controls (Figure 5E). In conclusion, RP11-521C20.3 inhibited apoptosis, promoted proliferation, and alleviated mitochondrial damage in CSE-treated A549 cells.
Discussion
COPD is a progressive inflammatory disease of the lung and is primarily caused by smoking.35,36 The effects of smoking go far beyond this, as it can also affect bone disorders.37 Apoptosis of lung structural cells is considered to be one of the pathogenic mechanisms of COPD.5,6 Our results showed that the number of apoptotic cells in the lungs of COPD rats was significantly increased when exposed to CS, which was accompanied by an increase in BMF protein levels. BMF is a member of the family of pro-apoptotic BH3-only proteins,14 indicating a close association with the apoptosis of lung structural cells in COPD.
BMF plays a regulatory role in apoptosis in a variety of diseases, such as cancer, neurodegenerative diseases, and ischemic stroke.38–40 FOXO3-activated BMF promotes apoptosis and inhibits tumor growth and metastasis in E-calcineurin-negative breast cancer cells.41 Increased BMF expression induced by STARD13 3′ UTR (a competing endogenous RNA) enhances BMF/Bcl-2 interaction in breast cancer cells, leading to the release of Bax and promoting apoptosis.38 Consistent with these findings, the present study revealed that overexpression of BMF promoted apoptosis. In human glioblastoma cells, Gas5 targets miR-222 and increases BMF expression, which inhibits Bcl-2 and upregulates Bax to promote apoptosis.42 In oxygen glucose deprivation-treated primary cortical neurons, lncRNA SHNG14 positively regulates BMF expression by sponging miR-181c-5p, which increases protein levels of Bax, cleaved caspase-3 to promote apoptosis.40 In the presence of Adriamycin, upregulated RBMS2 in breast cancer cells promotes BMF expression and increases expression of cleaved caspase-3, cleaved caspase-9, and poly ADP-ribose polymerase to exacerbate apoptosis.43 In addition, knockdown of Eomes in human colon cancer cells increases BMF expression, which is accompanied by increased levels of cleaved caspase-3 and cleaved caspase-7, and decreased expression of survivin, leading to apoptosis.44 In fact, our research showed that overexpression of BMF in CSE-treated A549 cells increased the expression of pro-apoptotic proteins (p53, caspase-3, and caspase-7) and inhibited the expression of anti-apoptotic proteins (Bcl-2 and survivin) to exacerbate apoptosis.
It was shown that RP11-521C20.3 was antisense to BMF mRNA,19 while natural antisense transcripts can regulate the expression of sense mRNA through several mechanisms.31 Our experiments confirmed that RP11-521C20.3 inhibited the expression of BMF mRNA and protein in CSE-treated A549 cells.
Some lncRNAs participate in the pathogenesis of COPD. For example, lncRNA SNHG5 and lncRNA MEG3 affect cellular proliferation and apoptosis, and tissue inflammation in the lung.26–29 Overexpression of lncRNA SHNG5 attenuates the effects of CSE on proliferation, apoptosis, and inflammation in 16HBE cells.26 Overexpression of lncRNA CASC2 attenuates apoptosis, inflammation in CSE-treated 16HBE cells.45 Furthermore, overexpression of lncRNA LOC729178 increases the level of Bcl-2 protein, decreased the levels of Bax, cleaved caspase-3 protein, and reduced the expression levels of inflammatory factors in CSE-treated 16HBE cells, ultimately alleviating CSE-induced apoptosis and inflammation.46 Similar to these findings, our datas suggested RP11-521C20.3 protected cells from CSE-induced apoptosis by regulating the expression of pro-apoptotic proteins (p53, caspase-3, and caspase-7) and anti-apoptotic proteins (Bcl-2 and survivin).
In conclusion, the above findings preliminarily confirmed our hypothesis. That is, RP11-521C20.3 negatively regulated the expression of BMF and played an important role in CSE-induced apoptosis of A549 cells.
Conclusion
Our study demonstrated that BMF exacerbated apoptosis in CSE-treated A549 cells by regulating the expression of its downstream pro- and anti-apoptotic molecules. RP11-521C20.3 may protect against CSE-induced apoptosis in A549 cells by targeting the BMF apoptosis signaling axis. These findings provide some insights into the pathogenesis of COPD and the development of BMF as an anti-apoptotic therapeutic target for COPD.
Abbreviations
BMF, Bcl-2 modifying factor; CSE, cigarette smoke extract; COPD, Chronic obstructive pulmonary disease; CS, Cigarette smoke; lncRNAs, Long non-coding RNAs; HE, hematoxylin and eosin; RT, room temperature; shRNA, short hairpin RNA.
Data Sharing Declaration
Upon request, data supporting the results of this manuscript are available from the corresponding author.
Ethics Approval
The study was approved by the Animal Experimentation Ethics Inspection Committee of Kunming Medical University (SCXK Dian2005-0003), and experimental animals received welfare in accordance with the National Institutes of Health guidelines.
Acknowledgments
The authors would like to thank TopEdit for its linguistic assistance during the preparation of this manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that was in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC, Grants 81860013).
Disclosure
The authors declare that they have no potential conflict of interest in this work.
References
1. GOLD. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease 2022 report. Available from: https://goldcopd.org/2022-gold-reports-2/.
2. Li X, Cao X, Guo M, Xie M, Liu X. Trends and risk factors of mortality and disability adjusted life years for chronic respiratory diseases from 1990 to 2017: systematic analysis for the Global Burden of Disease Study 2017. BMJ. 2020;368:m234. doi:10.1136/bmj.m234
3. Zhu B, Wang Y, Ming J, Chen W, Zhang L. Disease burden of COPD in China: a systematic review. Int J Chron Obstruct Pulmon Dis. 2018;13:1353–1364. doi:10.2147/copd.S161555
4. Agustí A, Hogg JC. Update on the Pathogenesis of Chronic Obstructive Pulmonary Disease. N Engl J Med. 2019;381(13):1248–1256. doi:10.1056/NEJMra1900475
5. Demedts IK, Demoor T, Bracke KR, Joos GF, Brusselle GG. Role of apoptosis in the pathogenesis of COPD and pulmonary emphysema. Respir Res. 2006;7(1):53. doi:10.1186/1465-9921-7-53
6. Song Q, Chen P, Liu XM. The role of cigarette smoke-induced pulmonary vascular endothelial cell apoptosis in COPD. Respir Res. 2021;22(1):39. doi:10.1186/s12931-021-01630-1
7. Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9(3):231–241. doi:10.1038/nrm2312
8. Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15(1):49–63. doi:10.1038/nrm3722
9. Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281(5381):1305–1308. doi:10.1126/science.281.5381.1305
10. Cheung CHA, Chang YC, Lin TY, Cheng SM, Leung E. Anti-apoptotic proteins in the autophagic world: an update on functions of XIAP, Survivin, and BRUCE. J Biomed Sci. 2020;27(1):31. doi:10.1186/s12929-020-0627-5
11. Kesavardhana S, Malireddi RKS, Kanneganti TD. Caspases in Cell Death, Inflammation, and Pyroptosis. Annu Rev Immunol. 2020;38:567–595. doi:10.1146/annurev-immunol-073119-095439
12. Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787(5):414–420. doi:10.1016/j.bbabio.2008.10.005
13. Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458(7242):1127–1130. doi:10.1038/nature07986
14. Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18(4):157–164. doi:10.1016/j.tcb.2008.01.007
15. Puthalakath H, Villunger A, O’Reilly LA, et al. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293(5536):1829–1832. doi:10.1126/science.1062257
16. Delgado M, Tesfaigzi Y. BH3-only proteins, Bmf and Bim, in autophagy. Cell Cycle. 2013;12(22):3453–3454. doi:10.4161/cc.26696
17. Delgado M, Tesfaigzi Y. Is BMF central for anoikis and autophagy? Autophagy. 2014;10(1):168–169. doi:10.4161/auto.26759
18. Du H, Wolf J, Schafer B, Moldoveanu T, Chipuk JE, Kuwana T. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem. 2011;286(1):491–501. doi:10.1074/jbc.M110.167148
19. Li C, Liu H, Zhang J, et al. LncRNA BMF-AS1 Exerts Anti-Apoptosis Function in COPD by Regulating BMF Expression. Pak J Zool. 2020;52(3):893–900. doi:10.17582/journal.pjz/20190909090912
20. Happo L, Strasser A, Cory S. BH3-only proteins in apoptosis at a glance. J Cell Sci. 2012;125(Pt5):1081–1087. doi:10.1242/jcs.090514
21. Piñon JD, Labi V, Egle A, Villunger A. Bim and Bmf in tissue homeostasis and malignant disease. Oncogene. 2008;27(Suppl1):S41–52. doi:10.1038/onc.2009.42
22. Strasser A. The role of BH3-only proteins in the immune system. Nat Rev Immunol. 2005;5(3):189–200. doi:10.1038/nri1568
23. Zhang H, Sun D, Li D, et al. Long non-coding RNA expression patterns in lung tissues of chronic cigarette smoke induced COPD mouse model. Sci Rep. 2018;8(1):7609. doi:10.1038/s41598-018-25702-3
24. Devadoss D, Long C, Langley RJ, et al. Long Noncoding Transcriptome in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol. 2019;61(6):678–688. doi:10.1165/rcmb.2019-0184TR
25. Pagano S, Negri P, Coniglio M, et al. Heat-not-burn tobacco (IQOS), oral fibroblasts and keratinocytes: cytotoxicity, morphological analysis, apoptosis and cellular cycle. An in vitro study. J Periodontal Res. 2021;56(5):917–928. doi:10.1111/jre.12888
26. Shen Q, Zheng J, Wang X, Hu W, Jiang Y, Jiang Y. LncRNA SNHG5 regulates cell apoptosis and inflammation by miR-132/PTEN axis in COPD. Biomed Pharmacother. 2020;126:110016. doi:10.1016/j.biopha.2020.110016
27. Lei Z, Guo H, Zou S, Jiang J, Kui Y, Song J. Long non-coding RNA maternally expressed gene regulates cigarette smoke extract induced lung inflammation and human bronchial epithelial apoptosis via miR-149-3p. Exp Ther Med. 2021;21(1):60. doi:10.3892/etm.2020.9492
28. Fan S, Ren Y, Zhang W, Zhang H, Wang C. Long non-coding maternally expressed gene 3 regulates cigarette smoke extract-induced apoptosis, inflammation and cytotoxicity by sponging miR-181a-2-3p in 16HBE cells. Oncol Lett. 2021;21(1):45. doi:10.3892/ol.2020.12306
29. Song B, Ye L, Wu S, Jing Z. Long non-coding RNA MEG3 regulates CSE-induced apoptosis and inflammation via regulating miR-218 in 16HBE cells. Biochem Biophys Res Commun. 2020;521(2):368–374. doi:10.1016/j.bbrc.2019.10.135
30. Latgé G, Poulet C, Bours V, Josse C, Jerusalem G. Natural Antisense Transcripts: molecular Mechanisms and Implications in Breast Cancers. Int J Mol Sci. 2018;19(1). doi:10.3390/ijms19010123
31. Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10(9):637–643. doi:10.1038/nrm2738
32. Wright JL, Churg A. Animal models of cigarette smoke-induced COPD. Chest. 2002;122(6Suppl):301s–306s. doi:10.1378/chest.122.6_suppl.301s
33. Lee JH, Lee DS, Kim EK, et al. Simvastatin inhibits cigarette smoking-induced emphysema and pulmonary hypertension in rat lungs. Am J Respir Crit Care Med. 2005;172(8):987–993. doi:10.1164/rccm.200501-041OC
34. Zhou AY, Zhao YY, Zhou ZJ, et al. Microarray Analysis of Long Non-Coding RNAs in Lung Tissues of Patients with COPD and HOXA-AS2 Promotes HPMECs Proliferation via Notch1. Int J Chron Obstruct Pulmon Dis. 2020;15:2449–2460. doi:10.2147/copd.S259601
35. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. 2017;389(10082):1931–1940. doi:10.1016/s0140-6736(17)31222-9
36. Mannino DM, Buist AS. Global burden of COPD: risk factors, prevalence, and future trends. Lancet. 2007;370(9589):765–773. doi:10.1016/s0140-6736(07)61380-4
37. Giuca MR, Pasini M, Drago S, et al. Influence of Vertical Facial Growth Pattern on Herbst Appliance Effects in Prepubertal Patients: a Retrospective Controlled Study. Int J Dent. 2020;2020:1018793. doi:10.1155/2020/1018793
38. Guo X, Xiang C, Zhang Z, Zhang F, Xi T, Zheng L. Displacement of Bax by BMF Mediates STARD13 3’UTR-Induced Breast Cancer Cells Apoptosis in an miRNA-Dependent Manner. Mol Pharm. 2018;15(1):63–71. doi:10.1021/acs.molpharmaceut.7b00727
39. Akhter R, Saleem S, Saha A, Biswas SC. The pro-apoptotic protein Bmf co-operates with Bim and Puma in neuron death induced by β-amyloid or NGF deprivation. Mol Cell Neurosci. 2018;88:249–257. doi:10.1016/j.mcn.2018.02.011
40. Bu X, Zhao Y, Chang M, Ge X. Downregulation of lncRNA SNHG14 alleviates neurons injury by modulating the miR-181c-5p/BMF axis in ischemic stroke. Brain Res Bull. 2021;174:379–388. doi:10.1016/j.brainresbull.2021.06.026
41. Hornsveld M, Tenhagen M, van de Ven RA, et al. Restraining FOXO3-dependent transcriptional BMF activation underpins tumour growth and metastasis of E-cadherin-negative breast cancer. Cell Death Differ. 2016;23(9):1483–1492. doi:10.1038/cdd.2016.33
42. Zhao X, Wang P, Liu J, et al. Gas5 Exerts Tumor-suppressive Functions in Human Glioma Cells by Targeting miR-222. Mol Ther. 2015;23(12):1899–1911. doi:10.1038/mt.2015.170
43. Xu F, Xia T, Xu QT, et al. RBMS2 Chemosensitizes Breast Cancer Cells to Doxorubicin by Regulating BMF Expression. Int J Biol Sci. 2022;18(4):1724–1736. doi:10.7150/ijbs.66480
44. Wang R, Kang Y, Löhr CV, et al. Reciprocal regulation of BMF and BIRC5 (Survivin) linked to Eomes overexpression in colorectal cancer. Cancer Lett. 2016;381(2):341–348. doi:10.1016/j.canlet.2016.08.008
45. Liu P, Zhang H, Zeng H, et al. LncRNA CASC2 is involved in the development of chronic obstructive pulmonary disease via targeting miR-18a-5p/IGF1 axis. Ther Adv Respir Dis. 2021;15:17534666211028072. doi:10.1177/17534666211028072
46. Wang M, Liu Y, Zhang Y, Zhang L. LncRNA LOC729178 acts as a sponge of miR-144-3p to mitigate cigarette smoke extract-induced inflammatory injury via regulating PHLPP2 in 16HBE cells. J Mol Histol. 2021;52(3):437–447. doi:10.1007/s10735-021-09972-2
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.