Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA RNF144A-AS1 Promotes Bladder Cancer Progression via RNF144A-AS1/miR-455-5p/SOX11 Axis
Authors Bi H, Shang Z, Jia C, Wu J, Cui B, Wang Q, Ou T
Received 1 July 2020
Accepted for publication 23 September 2020
Published 4 November 2020 Volume 2020:13 Pages 11277—11288
DOI https://doi.org/10.2147/OTT.S266067
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
This paper has been retracted.
Huifeng Bi,1,2 Zhenhua Shang,1 Chunsong Jia,1 Jiangtao Wu,1 Bo Cui,1 Qi Wang,1 Tongwen Ou1
1Department of Urology, Xuanwu Hospital Capital Medical University, Beijing, People’s Republic of China; 2Department of Urology, Jincheng General Hospital, Jincheng, Shanxi Province, People’s Republic of China
Correspondence: Tongwen Ou
Department of Urology, Xuanwu Hospital Capital Medical University, No. 45 Changchun Street, Xicheng District, Beijing 100053, People’s Republic of China
Tel +86 135 0106 5134
Fax +86 010 8319 8388
Email [email protected]
Background: Bladder cancer (BC) is the most commonly occurring malignant tumor of the urinary system worldwide. Long non-coding RNAs (lncRNAs), including lncRNA RNF144A-AS1 (RNF144A-AS1), perform an oncogenic role in BC progression. However, how RNF144A-AS1 is regulated in BC has not been fully investigated, and its role in BC is mostly obscure. In this study, we explore its role in BC progression.
Materials and Methods: The expression level of RNF144A-AS1 in BC tissues was explored via bioinformatics analysis and quantitative real-time PCR (qRT-PCR). We used RNF144A-AS1 siRNA (si-RNF144A-AS1) to inhibit the RNF144A-AS1 level in BC cell lines (J82 and 5637 cells). A series of experimental studies in vitro (CCK-8 assay, colony formation assay and Transwell assay) was performed to explore the role of si-RNF144A-AS1 on the proliferation, migration and invasion of J82 and 5637 cells. A BC xenograft model was established, and the effect of si-RNF144A-AS1 on xenograft growth was explored in vivo. The interactions among RNF144A-AS1, miR-455-5p and SOX11 were predicted by bioinformatics miRanda and Targetscan database, and verified by the luciferase reporter assay and RNA pull-down assay. Finally, miR-455-5p inhibitor and si-RNF144A-AS1 were cotransfected into J82 and 5637 cells.
Results: RNF144A-AS1 is overexpressed in BC tumors and cells, and its overexpression is correlated with poor prognosis. Knockdown of RNF144A-AS1 markedly suppressed the proliferation, migration and invasion of J82 and 5637 cells and significantly inhibited xenograft growth in nude mice, compared to si-NC. We found that RNF144A-AS1 serves as a sponge for miR-455-5p. Furthermore, a binding site of miR-455-5p was found in 3ʹ UTR of SOX11 gene, and overexpression of miR-455-5p suppressed SOX11 levels. RNF144A-AS1 knockdown markedly decreased SOX11 expression levels, while miR-455-5p inhibitor restored this repressive effect. Restoration of SOX11 could reverse this repressive effect of RNF144A-AS1 on cell proliferation, migration and invasion abilities.
Conclusion: Overall, our findings underline the critical role of RNF144A-AS1 in BC development, and our study reveals for the first time that RNF144A-AS1 promotes BC progression via the RNF144A-AS1/miR-455-5p/SOX11 axis.
Keywords: bladder cancer, lncRNA RNF144A-AS1, miRNA-455-5p, SOX11 gene, bladder cancer progression
Introduction
Bladder cancer (BC) is one of the most commonly occurring malignant tumors of the urinary system worldwide and was the eighth leading cause of cancer-related death in men in 2019.1 The current cancer treatment options for BC include conventional surgery, radiation therapy, chemotherapy and immunotherapy.2 These strategies have moderate effects on the early stages of BC, whereas it is difficult to treat the advanced stages of BC and the prognosis is very poor.3,4 Therefore, there is an urgent need to explore novel and effective molecular therapeutic targets for BC treatment.
Long non-coding RNAs (lncRNAs) are non-coding RNA transcripts5 that have been validated to play important roles in the development of cancers, including BC.6,7 Several lncRNAs are involved in the regulation of BC progression, such as lncRNA miR143HG,8 lncRNA DILC9 and lncRNA HOXA-AS27. From previously published data, lncRNA RNF144A-AS1 (RNF144A-AS1) was downregulated and promoted sensitivity and specificity in predicting chemoresistance in high-grade serous ovarian carcinoma cells.10 RNF144A-AS1 was also identified in glioblastoma multiforme and served as a potential lncRNA biomarker.11 However, only a few studies have reported its role in BC progression. One report suggested RNF144A-AS1 as a prognostic factor for BC using the least absolute shrinkage and selection operation Cox regression.12 Another study reported that RNF144A-AS1 was correlated with prognostic prediction for BC patients, and RNF144A-AS1 promoted the migration and invasion of BC cells in vitro.13 But the mechanism behind this promotion remains largely unclear.
In this study, we first explored the expression levels of RNF144A-AS1 in BC tumors or cell lines. We further investigated the critical role of RNF144A-AS1 in BC cell proliferation, migration and invasion by functional assays. BC xenografts were developed in nude mice. Moreover, we used bioinformatics analysis to predict the target miRNA of RNF144A-AS1 and investigated an lncRNA/miRNA/mRNA axis. We explored the role of this axis in BC progression and investigated the underlying mechanism. Overall, we aimed to provide a novel therapeutic target for BC treatment.
Materials and Methods
Clinical Samples
A total of 30 patients with BC were selected from Xuanwu Hospital Capital Medical University between 2017 and 2019. We collected BC tumor and the adjacent healthy tissues from the 30 subjects. All participants in this study provided informed written consent. The experiments were carried out according to the principles of Xuanwu Hospital Capital Medical University and approved by Xuanwu Hospital Capital Medical University Ethics Committee. The clinical characteristics of the patients are shown in Table 1.
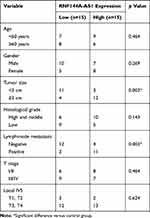 |
Table 1 Clinicopathological Features of the 30 Patients |
Cell Culture and Transfection
The normal bladder cell line (SV-HU-1) and five BC cell lines (RT4, 5637, J82, UMUC3 and T24) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) at 37°C in a 5% CO2 humidified incubator. Cell were plated into 96-well plates at a density of 3000 cells/well. The RNF144A-AS1 si-RNA (si-RNF144A-AS1) and its corresponding scrambled siRNA control (si-NC), and miR-455-5p and its nonsense control (miR-NC) plasmids were obtained from Thermo Fisher Scientific (Waltham, MA, USA). The miR-455-5p inhibitor (miR-455-5p inh) was obtained from Biomics (Nantong, Jiangsu, China). The plasmids were separately transfected into cells using a Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA).
Animal Experiments and Xenograft Collection
The nude mice were purchased from Charles River Laboratories (Beijing, China) and randomized into two groups (30 mice per group). The transfected si-NC or si-RNF144A-AS1 5637 cells (5×106 cells) were injected subcutaneously into nude mice. Tumor volumes and weights were measured at 7, 14, 28, 35 and 42 days after injection. All experiments were approved by the Animal Ethics Committee of Xuanwu Hospital Capital Medical University and were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Ministry of Health, China.
Total RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Valencia, CA, USA). qRT-PCR was performed to detect the expression levels of RNF144A-AS1, miR-455-5p and sex-determining region Y (SRY)-box protein 11 (SOX11). The expression level of miR-455-5p was detected using an miRNA real-time PCR assay kit (Aidlab, China) and the expression levels of RNF144A-AS1 and SOX11 were detected using Arraystar SYBR® Green Real-time qPCR Master Mix (Shanghai, China). U6 acted as the internal control for miRNA, while GAPDH acted as the internal control for lncRNA and mRNA. The primers used in this study were designed by Primer 5.0 and are listed in Table 2.
 |
Table 2 Primer List for qRT-PCR |
Western Blot
The proteins were extracted from BC cells by RIPA buffer (Beyotime, Beijing, China) and quantified with a Protein Quantification kit (Millipore, Billerica, MA, USA). The cell extracts were subjected to Western blot to determine the protein expression of SOX11 and Ago2. GAPDH was used as the protein loading control. Equal amounts of protein extracts were loaded on to SDS-PAGE gel and transferred to the PCDF membrane. After transfer, the membrane was blocked with 5% skim milk, followed by incubation with primary antibodies, SOX11 antibody (1:1000; Abcam, Shanghai, China), Ago2 antibody (1:1000; Abcam, Shanghai, China) or GAPDH antibody (1:1000; Santa Cruz, Santa Cruz, CA, USA). Subsequently, the membrane was washed and incubated with goat anti-rabbit IgG H&L (HRP) secondary antibody (1:2000; Abcam, Shanghai, China). ECL Reagent (Cell Signaling Technology, Danvers, MA, USA) was used for visualization and detection.
Cell Proliferation Assay (CCK-8 Assay)
For the cell proliferation assay, J82 and 5637 cells were transfected with si-NC or si-RNF144A-AS1, or cotransfected with si-RNF144A-AS1 and SOX11 plasmids. The transfected cells were seeded in 96-well plates (5×103 cells/well) and treated with 10 μL of CCK-8 solution (Beyotime, Shanghai, China). After incubation for 2 h at 37°C, optical density (OD) values were read at 450 nm by a microplate reader (Bio-Rad, Hercules, CA, USA).
Dual-Luciferase Reporter Assay
The wide type (wt) or mutant (mut) RNF144A-AS1 and SOX11 were synthesized by Shanghai GenePharma Co. and cloned into pmirGLO luciferase vector (Promega, Madison, WI, USA). For the reporter assay, cells were cotransfected with reporter vector and miR-455-5p mimics or NC mimics using Lipofectamine 2000 (Invitrogen, San Diego, CA, USA). Following a 48-h transfection period, the luciferase activities were measured using a dual luciferase reporter assay system (Promega, Madison, WI, USA).
Pull-Down Assay
For the pull-down assay, J82 and 5637 cells were transfected with 20 nM of biotinylated miR-NC (bio-NC) or biotinylated miR-455-5p (bio-miR-455-5p). After transfection for 48 h, cells lysates were obtained by sonication and incubated with Pierce™ Streptavidin Magnetic Beads (Thermo Fisher Scientific, Waltham, MA, USA). Lysates were purified by the RNeasy Mini Kit (Qiagen, Duesseldorf, Germany), following detection of RNF144A-AS1 enrichment by qRT-PCR and Western blot. During this experiment, 10% lysates served as the input control.
Nuclear and Cytoplasmic RNA Extraction
For nuclear and cytoplasmic RNA isolation, the PARISTM Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used according the manufacturer’s protocol. The nuclear and cytoplasmic levels of RNF144A-AS were detected by qRT-PCR.
Transwell Assay
The invasion ability of bladder cells was determined using a Transwell chamber (Corning, NY, USA). In brief, cells were digested with trypsin and resuspended with serum-free medium. A total of 1×104 cells in 200 µL of serum-free medium were added into the upper chamber, which was precoated with Matrigel (BD, Franklin Lakes, USA), and 800 µL DMEM with 30% FBS was added to the lower chamber. The chamber was incubated at 37°C for 24 h. The transmigrated cells were fixed with paraformaldehyde (Sinopharm Chemical Reagent Co., Shanghai, China) and stained with crystal violet (Solarbio, Beijing, China). The stained cells were photographed and counted with an inverted microscope (Motic China Group Co., Xiamen, China). The migration ability was investigated by a similar method, except that the upper chamber was not precoated with Matrigel.
Statistical Analysis
All statistical analysis was performed with GraphPad Prism 6 software. The survival curves were assessed by Kaplan–Meier analysis and the significance of the clinicopathological parameters of BC patients was determined by the chi-squared test. For the functional assays in vitro, the difference between two groups was analyzed by the Student’s t-test and differences among multiple groups were analyzed by ANOVA with Bonferroni correction. p<0.05 was considered statistically significant.
Results
RNF144A-AS1 is Significantly Upregulated in BC and Associated with Poor Prognosis
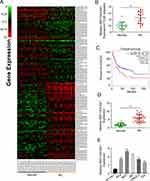
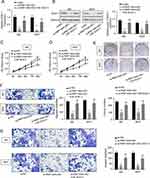
First, 19 BC patients from TCGA database were included and we compared the RNF144A-AS1 expression levels of BC tumor samples with adjacent normal tissues. The cluster heatmap showed that RNF144A-AS1 was highly expressed in BC tumors compared to adjacent normal tissues (Figure 1A and B). Through Gene Expression Profiling Interactive Analysis (GEIPA), we found that a high level of RNF144A-AS1 in BC was correlated with poor prognosis (Figure 1C).
We also evaluated the level of RNF144A-AS1 in BC tumor samples (n=30) and BC cell lines by qRT-PCR. The results showed that RNF144A-AS1 was significantly upregulated in BC tumor samples and five BC cell lines compared to adjacent normal tissues (p<0.001) and SV-HU-1 cells (p<0.01) (Figure 1D and E). As shown in Figure 1E, the levels of RNF144A-AS1 in J82 and 5637 cells were higher than in other BC cells. Hence, J82 and 5637 cell lines were chosen for further experiments in this study.
Silencing of RNF144A-AS1 Inhibits BC Cell Proliferation, Migration and Invasion
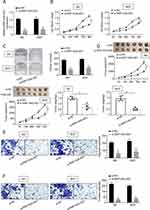
To determine whether RNF144A-AS1 silencing played a critical role in BC cell proliferation, migration and invasion, the J82 and 5637 cells were transfected with RNF144A-AS1 siRNA (si-RNF144A-AS1) or its corresponding scrambled siRNA control (si-NC). qRT-PCR results indicated that si-RNF144A-AS1 effectively suppressed the level of RNF144A-AS1 in J82 and 5637 cells (Figure 2A).
The CCK-8 assay demonstrated that knockdown of RNF144A-AS1 significantly decreased the capacity for cell proliferation in J82 and 5637 cells (Figure 2B). Colony formation assays revealed that interference of RNF144A-AS1 significantly suppressed colony formation (Figure 2C). Through subcutaneous tumor formation assays, we found that si-RNF144A-AS1 significantly inhibited xenograft volume and weight in nude mice, compared to si-NC (Figure 2D). The Transwell assay indicated that knockdown of RNF144A-AS1 significantly reduced the cell migration and invasion abilities of J82 and 5637 cells (Figure 2E and F).
RNF144A-AS1 Directly Interacts with miR-455-5p in BC Cells
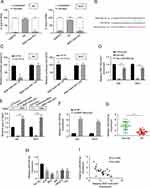
We detected the level of RNF144A-AS1 in the cytoplasm and nucleus of BC cells. The qRT-PCR results showed that RNF144A-AS1 was mainly located in the cytoplasm of J82 and 5637 cells (Figure 3A).
We employed starBase software to nominate the putative targets of RNF144A-AS1. We found that the 3ʹ UTR fragment of human miR-455-5p contains an RNF144A-AS1-binding site (Figure 3B). The dual-luciferase reporter assay showed that miR-455-5p mimics significantly reduced the luciferase activity of RNF144A-AS1 wt, but did not impact the luciferase activity of RNF144A-AS1 mut (Figure 3C). To further investigate the interaction between RNF144A-AS1 and miR-455-5p, an RNA pull-down assay was performed, which found RNF144A-AS1 enrichment in bio-miR-455-5p beads, but no enrichment in bio-NC beads (Figure 3D and E). Moreover, we found that knockdown of RNF144A-AS1 significantly increased the expression level of miR-455-5p in J82 and 5637 cells (Figure 3F).
Meanwhile, the expression level of miR-455-5p was markedly downregulated in BC tumor tissues compared to the adjacent normal tissues (Figure 3G). In several BC cell lines, the expression level of miR-455-5p was markedly suppressed compared to SV-HU-1 cells (Figure 3H). Spearman correlation statistical analysis illustrated that the expression levels of RNF144A-AS1 and miR-455-5p were negatively correlated in BC tumor tissues (Figure 3I). These results indicated that there was a putative RNF144A-AS1 binding site in the 3ʹ UTR of miR-455-5p.
RNF144A-AS1 Promotes SOX11 Expression by Sponging miR-455-5p
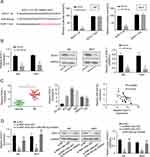
Through Targetscan software, we found an miR-455-5p binding site in the 3ʹ UTR fragment of the SOX11 gene (Figure 4A). As shown in Figure 4A, the dual-luciferase reporter assay showed that miR-455-5p mimics significantly reduced the luciferase activity of SOX11 wt, but did not impact the luciferase activity of SOX11 mut. The mRNA and protein expression levels of SOX11 were shown to be downregulated in miR-455-5p-overexpressed J82 and 5637 cells (Figure 4B). The expression level of SOX11 was significantly upregulated in BC tumor tissues and cell lines compared to the adjacent normal tissues or SV-HU-1 cells (Figure 4C). Spearman correlation statistical analysis illustrated that the expression levels of RNF144A-AS1 and miR-455-5p were negatively correlated in BC tumor tissues (Figure 4C).
Moreover, to prove the correlation between RNF144A-AS1 and SOX11, qRT-PCR and Western blot were performed to detect the level of SOX11 in RNF144A-AS1 silencing cells or cotransfected si-RNF144A-AS1 and miR-455-5p inh cells. We found that RNF144A-AS1 knockdown markedly decreased SOX11 expression levels, while miR-455-5p inh restored the repressive effect of RNF144A-AS1 silencing on SOX11 expression levels in J82 and 5637 cells (Figure 4D).
Restoration of SOX11 Reverses the Effects of RNF144A-AS1 Knockdown
To explore whether RNF144A-AS1 performs functions via regulating SOX11, we transfected SOX11 plasmid (overexpression of SOX11, OE SOX11) into RNF144A-AS1 silencing cells. We found that RNF144A-AS1 knockdown markedly decreased SOX11 expression levels, while OE SOX11 restored the repressive effect of RNF144A-AS1 silencing (p<0.05) (Figure 5A and B).
Furthermore, our results showed that OE SOX11 significantly restored the repressive effect of RNF144A-AS1 silencing on cell proliferation (CCK-8 assay, colony formation assay; Figure 5C–E), migration (Transwell assay; Figure 5F) and invasion (Transwell assay; Figure 5G) of J82 and 5637 cells.
Discussion
The rate of BC patients with lymphatic metastasis is high and it has been shown to be a significant obstacle for BC treatment.14 To date, the treatment of choice for BC is combined surgery, chemotherapy, radiotherapy and immunotherapy, which has achieved recent advances. Unfortunately, the 5-year survival rate gradually declines with BC progression, falling to less than 50% at later stages.15 Hence, there is a need to find novel, effective, reliable biomarkers that act as molecular therapeutic targets or prognostic factors for BC.
LncRNAs are non-protein-coding transcripts longer than 200 nucleotides.16–18 A vast number of lncRNAs has been discovered and been evidenced to exert crucial functions in biological processes, including cell proliferation, migration and invasion.19–21 Meanwhile, meta-analysis of several studies revealed that the expression levels of lncRNAs are associated with the prognosis of BC.22
Increasing numbers of studies have reported that lncRNAs can serve as competing endogenous RNAs (ceRNAs) to bind miRNAs, consequently regulating the expression levels of downstream genes.23,24 Some lncRNAs have been identified as molecular biomarkers for BC patients; however, novel correlated lncRNAs are urgently needed.25–27
RNF144A-AS1 has been identified as an optimal diagnostic and prognostic biomarker for several cancer types,10−12−28, and has also been shown to promote cell migration and invasion in BC.13 However, the mechanism behind the regulation remains largely unclear.
In this study, we found that RNF144A-AS1 level was markedly upregulated in BC tumors and cell lines, and its high level in BC was correlated with poor prognosis. We further explored its involvement in BC progression in vivo and in vitro. Knockdown of RNF144A-AS1 in BC cells significantly suppressed cell proliferation, migration and invasion abilities, and has been shown to have an effect on xenograft volume and weight in nude mice as well. These findings are consistent with previous studies12,13 showing a strong association between RNF144A-AS1 and BC progression.
Notably, we explored the role of RNF144A-AS1 as a ceRNA in this study. miR-455-5p has been reported to serve as a tumor suppressor for several cancer types by regulating tumor development and differentiation.29–32 One previous study reported the critical role of miR-455-5p in BC progression, demonstrating that miR-455-5p suppressed BC cell migration, invasion and proliferation by directly targeting the 3ʹ UTR region of tight junction protein 1.32 Through bioinformatics analysis and functional assays, RNF144A-AS1 was first found to act as a sponge for miR-455-5p in our study. Thus, we concluded that RNF144A-AS1 performed its biological functions in BC progression by sponging miR-455-5p.
Furthermore, we found one binding site for miR-455-5p in the mRNA SOX11 3ʹ UTR region. SOX11 performs a variety of functions in biological processes, such as adult neurogenesis and tumorigenesis.33 SOX11 serves as a tumor suppressor against the development of several cancer types, such as ovarian cancer,34,35 mantle cell lymphoma36 and BC.37 Wu et al reported that the SOX11 level was significantly upregulated in BC tissues and was shown to promote the proliferation and apoptosis of BC cells.37
In our study, SOX11 expression was also demonstrated to be markedly upregulated in BC cells and its expression was regulated by RNF144A-AS1 by sponging miR-455-5p. RNF144A-AS1 knockdown markedly decreased SOX11 expression levels, while OE SOX11 restored the repressive effect of RNF144A-AS1 silencing. Moreover, OE SOX11 significantly restored the repressive effect of RNF144A-AS1 silencing on cell proliferation, migration and invasion of J82 and 5637 cells. We verified that RNF144A-AS1 functioned as a ceRNA for miR-455-5p in regulating SOX11, and this axis promoted the cell proliferation, migration and invasion of BC cells.
These findings revealed that RNF144A-AS1 is an oncogene for BC and it exerts promoting effects on cell proliferation, migration and invasion in BC tumor progression. This study first revealed the underlying mechanism behind this promotion, that RNF144A-AS1 drives BC progression by upregulating YAP via sponging miR-455-5p. This study provides a novel therapeutic target for BC treatment.
Ethics Approval
All experiments were approved by the Animal Ethics Committee of Xuanwu Hospital Capital Medical University and were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Ministry of Health, China.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed on the journal to which the article will be submitted; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (grant number ZYLX201801).
Disclosure
The authors declare that there are no competing interests associated with this manuscript.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Jiang QQ, Liu B, Yuan T. MicroRNA-16 inhibits bladder cancer proliferation by targeting Cyclin D1. Asian Pac J Cancer Prev. 2013;14(7):4127–4130. doi:10.7314/APJCP.2013.14.7.4127
3. Siefker-Radtke A, Curti B. Immunotherapy in metastatic urothelial carcinoma: focus on immune checkpoint inhibition. Nat Rev Urol. 2018;15(2):112–124.
4. Chen C, Luo Y, He W, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404–421. doi:10.1172/JCI130892
5. Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):5–7. doi:10.1038/nsmb.2942
6. Chen JB, Zhu YW, Guo X, et al. Microarray expression profiles analysis revealed lncRNA OXCT1-AS1 promoted bladder cancer cell aggressiveness via miR-455-5p/JAK1 signaling. J Cell Physiol. 2019;234(8):13592–13601. doi:10.1002/jcp.28037
7. Wang F, Wu D, Chen J, et al. Long non-coding RNA HOXA-AS2 promotes the migration, invasion and stemness of bladder cancer via regulating miR-125b/Smad2 axis. Exp Cell Res. 2019;375(1):1–10. doi:10.1016/j.yexcr.2018.11.005
8. Xie H, Huang H, Huang W, Xie Z, Yang Y, Wang F. LncRNA miR143HG suppresses bladder cancer development through inactivating Wnt/beta-catenin pathway by modulating miR-1275/AXIN2 axis. J Cell Physiol. 2019;234(7):11156–11164. doi:10.1002/jcp.27764
9. Ma QY, Li SY, Li XZ, et al. Long non-coding RNA DILC suppresses bladder cancer cells progression. Gene. 2019;710:193–201. doi:10.1016/j.gene.2019.06.009
10. Song J, Zhang W, Wang S, Liu K, Song F, Ran L. A panel of 7 prognosis-related long non-coding RNAs to improve platinum-based chemoresistance prediction in ovarian cancer. Int J Oncol. 2018;53(2):866–876.
11. Li Y, Guo D. Identification of novel lncRNA markers in glioblastoma multiforme and their clinical significance: a study based on multiple sequencing data. Onco Targets Ther. 2020;13:1087–1098. doi:10.2147/OTT.S235951
12. He A, He S, Peng D, et al. Prognostic value of long non-coding RNA signatures in bladder cancer. Aging (Albany NY). 2019;11(16):6237–6251. doi:10.18632/aging.102185
13. Wang Y, Du L, Yang X, et al. A nomogram combining long non-coding RNA expression profiles and clinical factors predicts survival in patients with bladder cancer. Aging (Albany NY). 2020;12(3):2857–2879. doi:10.18632/aging.102782
14. Mari A, Kimura S, Foerster B, et al. A systematic review and meta-analysis of lymphovascular invasion in patients treated with radical cystectomy for bladder cancer. Urol Oncol. 2018;36(6):293–305. doi:10.1016/j.urolonc.2018.03.018
15. Fonteyne V, Ost P, Bellmunt J, et al. Curative treatment for muscle invasive bladder cancer in elderly patients: a systematic review. Eur Urol. 2018;73(1):40–50. doi:10.1016/j.eururo.2017.03.019
16. Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30(10):439–452. doi:10.1016/j.tig.2014.08.004
17. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166.
18. Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–437.
19. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606
20. Jiang W, Xia J, Xie S, et al. Long non-coding RNAs as a determinant of cancer drug resistance: towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist Updat. 2020;50:100683. doi:10.1016/j.drup.2020.100683
21. Wu Y, Wang Y, Wei M, Han X, Xu T, Cui M. Advances in the study of exosomal lncRNAs in tumors and the selection of research methods. Biomed Pharmacother. 2020;123:109716. doi:10.1016/j.biopha.2019.109716
22. Zhong Y, Zhang Y, Li H, Ma W. LncRNAs act as prognostic biomarkers in bladder carcinoma: A meta-analysis. Heliyon. 2019;5(11):e02785. doi:10.1016/j.heliyon.2019.e02785
23. Zhao Z, Sun W, Guo Z, Zhang J, Yu H, Liu B. Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 2019;116900.
24. Wang JY, Yang Y, Ma Y, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:109627. doi:10.1016/j.biopha.2019.109627
25. Dyrskjot L, Reinert T, Algaba F, et al. Prognostic impact of a 12-gene progression score in non-muscle-invasive bladder cancer: a prospective multicentre validation study. Eur Urol. 2017;72(3):461–469. doi:10.1016/j.eururo.2017.05.040
26. Kluth LA, Black PC, Bochner BH, et al. Prognostic and prediction tools in bladder cancer: a comprehensive review of the literature. Eur Urol. 2015;68(2):238–253. doi:10.1016/j.eururo.2015.01.032
27. Zhang C, Li Z, Hu J, Qi F, Li X, Luo J. Identification of five long noncoding RNAs signature and risk score for prognosis of bladder urothelial carcinoma. J Cell Biochem. 2020;121(1):856–866. doi:10.1002/jcb.29330
28. Hu Y, Guo G, Li J, Chen J, Tan P. Screening key lncRNAs with diagnostic and prognostic value for head and neck squamous cell carcinoma based on machine learning and mRNA-lncRNA co-expression network analysis. Cancer Biomark. 2020;27(2):195–206. doi:10.3233/CBM-190694
29. Qin L, Zhang Y, Lin J, Shentu Y, Xie X. MicroRNA-455 regulates migration and invasion of human hepatocellular carcinoma by targeting Runx2. Oncol Rep. 2016;36(6):3325–3332. doi:10.3892/or.2016.5139
30. Liu J, Zhang J, Li Y, Wang L, Sui B, Dai D. MiR-455-5p acts as a novel tumor suppressor in gastric cancer by down-regulating RAB18. Gene. 2016;592(2):308–315. doi:10.1016/j.gene.2016.07.034
31. Chai J, Wang S, Han D, Dong W, Xie C, Guo H. MicroRNA-455 inhibits proliferation and invasion of colorectal cancer by targeting RAF proto-oncogene serine/threonine-protein kinase. Tumour Biol. 2015;36(2):1313–1321. doi:10.1007/s13277-014-2766-3
32. Tsai KW, Kuo WT, Jeng SY. Tight junction protein 1 dysfunction contributes to cell motility in bladder cancer. Anticancer Res. 2018;38(8):4607–4615. doi:10.21873/anticanres.12765
33. Oliemuller E, Kogata N, Bland P, et al. SOX11 promotes invasive growth and ductal carcinoma in situ progression. J Pathol. 2017;243(2):193–207. doi:10.1002/path.4939
34. Fang G, Liu J, Wang Q, et al. MicroRNA-223-3p regulates ovarian cancer cell proliferation and invasion by targeting SOX11 expression. Int J Mol Sci. 2017;18:6.
35. Sernbo S, Gustavsson E, Brennan DJ, et al. The tumour suppressor SOX11 is associated with improved survival among high grade epithelial ovarian cancers and is regulated by reversible promoter methylation. BMC Cancer. 2011;11:405.
36. Meggendorfer M, Kern W, Haferlach C, Haferlach T, Schnittger S. SOX11 overexpression is a specific marker for mantle cell lymphoma and correlates with t(11;14) translocation, CCND1 expression and an adverse prognosis. Leukemia. 2013;27(12):2388–2391. doi:10.1038/leu.2013.141
37. Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24(1):40. doi:10.1186/s10020-018-0039-0
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.