Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA RMRP Promotes Cell Proliferation and Invasion Through miR-613/NFAT5 Axis in Non-Small Cell Lung Cancer
Received 23 March 2020
Accepted for publication 11 August 2020
Published 8 September 2020 Volume 2020:13 Pages 8941—8950
DOI https://doi.org/10.2147/OTT.S255126
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Mingjun Yang, Honggang Ke, Wen Zhou
Department of Cardiothoracic Surgery, Affiliated Hospital of Nantong University, Nantong 226021, Jiangsu Province, People’s Republic of China
Correspondence: Wen Zhou
Department of Cardiothoracic Surgery, Affiliated Hospital of Nantong University, Nantong 226021, Jiangsu Province, People’s Republic of China
Email [email protected]
Background: The abnormal expression of RMRP and miR-613 was respectively associated with the pathogenesis of lung cancer, but the role of the RMRP/miR-613 axis in NSCLC has not been studied.
Methods: In this report, we measured the levels of RMRP in clinical NSCLC samples and cell lines. The target gene of RNA was predicted by online tools and verified by Luciferase reporter assay. Moreover, the function and regulatory mechanism of RMRP in the progression of cancer were further investigated.
Results: Our data showed that the expression of RMRP in NSCLC tissues and cell lines was both up-regulated. Functionally, RMRP promoted the proliferation and metastasis of A549 and H1299 cells. Luciferase reporter assay confirmed that RMRP was the sponger of miR-613, and NFAT5 is the direct target of miR-613. Functional acquisition and loss-of-function strategies further confirmed that RMRP induces the up-regulation of NFAT5 expression through competitive binding with miR-613, leading to promote the progression and metastasis potential of lung cancer cells.
Conclusion: Collectively, our findings emphasized the importance of RMRP in the development of NSCLC, which may provide a new therapeutic target and potential diagnostic biomarker for NSCLC therapy.
Keywords: NSCLC, RMRP, miR-613, NFAT5, sponge
Introduction
In the world, lung carcinoma ranks the NO. 1 cancer in the incidence and mortality of all the malignant tumors.1 According to WHO guidelines, lung carcinoma can be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), of which NSCLC accounts for about 80%.2 Although the significant progress based on the targeted molecular drugs prolongs the survival time of patients with advanced lung cancer, the overall long-term survival rate has not been bona fide improved.3 The main reason is that most NSCLC patients are diagnosed with advanced stage.4 Therefore, a more in-depth elucidation of the molecular biological mechanism in the occurrence and development of NSCLC is exceedingly pivotal for the early screening and treatment.
Long non-coding RNA (lncRNA) is one type of RNA more than 200 nt and no protein-coding function, participating in many essential regulatory processes.5 In recent years, accumulating evidence indicates that lncRNAs play essential roles in tumor progression and metastasis.6 Among them, the RNA component of mitochondrial RNA processing endoribonuclease (RMRP) is a lncRNA expressed in various tissues of mice and humans.7 To date, RMRP has been demonstrated to participate in the tumorigenesis of several cancers, such as hepatocellular carcinoma,8 bladder cancer,9 and papillary thyroid cancer.10 For NSCLC, the RMRP was up-regulated in clinical samples and served as a ceRNA for miR-1-3p to promote the progression of NSCLC.11 However, the miR-675/MAPK1 axis was found to be regulated by RMRP in papillary thyroid cancer, promoting the growth and metastasis of cancer cells.10 Another study revealed that miR-613 was a sponger of RMRP, leading to the tumorigenesis of hepatocellular carcinoma.8 Increasing studies have shown that miR-613 plays a critical regulatory role in a large number of cancers.12 Of note, miR-613 is also found to be down-regulated in lung cancer.13,14
In this report, we also found that the RMRP level was up-regulated in NSCLC clinical samples and cell lines. Overexpression of RMRP up-regulated the proliferation and metastasis potential of A549 and H1299 cells. Mechanismly, RMRP inhibited the expression of miR-613 to up-regulate its target NFAT5 expression. Moreover, miR-613 inhibitor reversed the proliferation and migration of NSCLC cells inhibited by RMRP silencing. These findings suggested that RMRP up-regulates cell proliferation and invasion/migration via the miR-613/NFAT5 axis in NSCLC, maybe providing a novel target and potential diagnostic marker for NSCLC therapy.
Materials and Methods
Clinical Specimens
Fresh NSCLC tissues were acquired from 80 patients at Affiliated Hospital of Nantong University from June 2017 to December 2019. And the healthy lung tissues at the edge of the lung tumor were obtained as the paracancerous tissue (control). Subsequently, the specimens were quickly snap-frozen in liquid nitrogen and transferred to −80°C for preservation. The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Animal Ethics Committee of Affiliated Hospital of Nantong University. All the patients signed informed consent, and the hospital ethical committee approved this study.
Cell Treatment
HCT116 (human colorectal cancer cell lines) and five types of NSCLC cells, including HCC827, 16HBE, H1299, H1975, and A549 cells, were obtained from ATCC and cultured according to their recommendations. miRNA mimics/inhibitors and their corresponding control were commercially obtained from GenePharm Co. Ltd. (Shanghai, China). For gene overexpression, the full-length gene of RMRP was cloned into the pcDNA3.1(+) vector (Sigma, MO, USA). In the experiment of gene knockdown, short hairpin RNA (shRNA) targeting the RMRP (5ʹ-CTG AGG ACG TGG GAT GAT CAT GGA CAT GAG GAT TAC CCA TGT ACT AGT AGA TCT CGT ACA CCA TCA GGG TAC GTT CCA GCT AGCT-3ʹ) and NFAT5 (5ʹ-CCG GGC AAT GGA AGT GTT GAC TTG GCT CGA GCC AAG TCA ACA CTT CCA TTG CTT TTTG-3ʹ) was commercially synthesized and cloned into pLKO.1 vector (Sigma, MO, USA). When the cell density reached 70–80%, plasmids, and miRNA mimics or inhibitor were transfected by Lipofectamine 2000 (ThermoFisher, Shanghai, China). To generate stable cell lines, transfected cells were selected with G418 (600 µg/mL) or puromycin (1 µg/mL; Sigma, MO, USA). Under the specified plural number of infection and treatment time, these vectors and miRNA mimics or inhibitor have neither side effects, including affecting the adherence, shape, or activity of lung cells mentioned above. The sequences were synthesized as followed: miR-613 mimic, 5ʹ -CAA GUG UGA AGG GAC CCU UCC-3ʹ; siRNA NC, 5ʹ -UUC UCC GAA CGU GUC ACG UTT-3ʹ; miR-613 inhibitor, 5ʹ-CCU UCC UGU AGU GUC UUAU-3ʹ and inhibitor NC, 5ʹ-GAT GGT CTT GCG GTC GTA GAT-3ʹ.
Animal Experiments
All animal experiments were conducted following the internationally accepted laboratory animal use and care guidelines15 and approved by the Animal Care and Utilization institutions Review Committee of Cancer Hospital of Affiliated Hospital of Nantong University. 6-week-old male Balb/c nude mice were placed in the absence of specific pathogen (SPF) and randomly divided into pLKO.1 group, pLKO.1-shRMRP group, pcDNA3.1(+) plasmids group, and pcDNA3.1-RMRP group (n=3 in each group). For tumorigenicity in vivo, 6×106 stably transfected A549 cells were subcutaneously injected into the right side of mice. The tumor volume was measured every week and calculated according to the formula: 0.5 × length × width.2 All mice were euthanatized 28 days later, and the tumors were surgically removed, measured, and stored in liquid nitrogen for further experiments.
Quantitative Real-Time PCR (qRT-PCR)
Cells were treated according to the experimental group. When the density reached 90%, the RNA extraction was performed by TRIzol reagent (Beyotime, Nantong, China). According to the instructions, the extracted total RNA was reverse transcribed to cDNA by PrimeScript 1st Strand cDNA synthesis kit (Takara, Otsu, Japan). After the implementation of real-time reverse-transcription polymerase chain reaction, the CT value of the target gene expression was obtained compared with the control group. GAPDH and U6 were used as the internal control gene. The relative quantitative analysis was carried out by 2–ΔΔCT method. The probes were synthesized as followed: GAPDH-R: 5ʹ-GAG TCC TTC CAC GAT ACC AA-3ʹ; miR-613-F: 5ʹ-GTG AGT GCG TTT CCA AGT GT-3ʹ; U6-F: 5ʹ-CCG CCC GCC GCC AGG CCCC-3ʹ; U6-R: 5ʹ-ATA TGG AAC GCT TCA CGA ATT-3ʹ; RMRP-F: 5ʹ-ACT CCA AAG TCC GCC AAGA-3ʹ; RMRP-R: 5ʹ-TGC GTA ACT AGA GGG AGC TGAC-3ʹ; GAPDH-F: 5ʹ-GGG AGC CAA AAG GGT CAT-3ʹ; miR-613-R: 5ʹ-TGA GTG GCA AAG AAG GAA CAT-3ʹ; NFAT5-F: 5ʹ-GAA GTG GAC ATT GAA GGC-3ʹ; NFAT5-F: 5ʹ-CTG GCT TCG ACA TCA GCA TT-3ʹ.
Western Blot Assay
The total protein was extracted and quantified by BCA assay (Beyotime, Nantong, China). Then, the lysate was analyzed by 12% SDS-PAGE, followed by transferring onto polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA, USA). After being blocked with 5% (w/v) non-fat dry milk, the membranes were incubated with antibodies, including anti-GAPDH (1:2000), anti-PCNA (1:1000), anti-Cox-2 (1:1000), anti-MMP2 (1:1000), and anti-MMP9 (1:1000) at 4°C, respectively. All the antibodies were purchased from Abcam (Cambridge, UK). Then, the appropriate HRP-conjugated secondary antibodies (1:5000, Proteintech, Wuhan, China) were applied for 1 hour at room temperature. The protein bands were detected by ECL luminescence kit (Pierce, Rockford, IL, USA). Quantitative analysis was carried out on ImageJ 1.8.
CCK-8 Assay
Cells were cultured in corresponding complete medium in 96-well plates. When the density reached 70%, cells were transfected as mention above. After 48 hours transfection, 10 μL CCK-8 solution (Beyotime, Nantong, China) was added to each well. After incubating for 60 minutes, the absorbance was detected by a microplate reader at 450nm wavelength (Thermo Fisher, Massachusetts, USA).
Colony Formation Assay
The transfected A549 and H1299 were seeded into a 6-well plate (5×103/well) at 37°C and cultured with 5% CO2. After 16 days, the cells were washed with phosphate buffer, fixed overnight with paraformaldehyde, and was stained with 0.4% crystal violet (Solarbio Science and Technology Ltd., Beijing, China) for 30 minutes. The colony counts were carried out under the inverted microscope.
Transwell Chamber Assay
For the transwell migration and invasion assay, a 24-well transwell chamber that the insert membranes were coated with or without diluted Matrigel was used. The transfected A549 and H1299 cells (1×105 cells) were added to the upper chamber and cultured for 24 hours. Subsequently, the insert membranes were cut and stained with crystal violet (Solarbio Science and Technology Ltd., Beijing, China). The images were taken by an inverted microscope, and the number of invading cells was counted in three wells per group.
Luciferase Reporter Gene Assay
The wild-type (WT) RMRP transcript containing miR-613 binding site and mutant (Mut) RMRP transcript with the mutated miR-613 binding site was cloned into the psiCHECK-2 vector (Promega, Beijing, China) to construct psiCHECK-2-RMRP-WT and psiCHECK-2-RMRP-Mut reporter plasmid. Moreover, the 3ʹ-UTRs (wild type) and the corresponding mutant 3ʹ-UTRs of NFAT5 were amplified by PCR. Obtained target fragments were cloned into the psiCHECK-2 reporter vector to construct psiCHECK-2-NFAT5-WT and psiCHECK-2-NFAT5-Mut reporter plasmid. A549 cells were inoculated into 24-well plate at the concentration of 1 × 105 cells/cm2. When the cell density reached 70–80%, reporter gene plasmids, RMRP overexpression plasmids, and miR-613 mimics or mimic control were co-transfected into A549 cells by Lipofectamine 2000 (ThermoFisher, Shanghai, China). The cells were collected 48 hours after transfection, and luciferase activity was detected.
Statistical Analysis
Data were presented with mean ± SD. Independent-samples T-tests assuming equal variance was used for statistical analysis, and statistical software SPSS17.0 was used. P < 0.05 was considered to be statistically significant. All experiments were independently repeated at least three times.
Results
High Expression of RMRP is Associated with the Pathogenesis of NSCLC
In order to explore the physiological expression of RMRP, the levels of RMRP in 80 matched lung cancer tissues and corresponding paracancerous tissues were measured by qRT-PCR. As a result, the level of RMRP in tumor tissues was higher than that in paracancerous tissues (P<0.05; Figure 1A). Besides, in these 80 cases of NSCLC, the expression levels of RMRP in 30 advanced cases (stage III and IV) was higher than that in 50 early-stage cases (stage I and II) (P<0.01; Figure 1B). At the cellular levels, compared with HCT116 cells, the expression levels of RMRP in HCC827, 16HBE, H1299, H1975, and A549 cells were significantly accordingly up-regulated (P<0.01; Figure 1C), indicating that up-regulation of RMRP is positively correlated with the lung cancer progression. Kaplan-Meier analysis revealed that overall survival rates of NSCLC patients with high RMRP levels were decreased compared to the low RMRP levels group (P<0.01; Figure 1D). These clinical data suggested that the overexpression of RMRP has adverse effects on the prognosis of NSCLC.
RMRP Promotes NSCLC Cell Proliferation and Metastasis Potential in vitro
Subsequently, we addressed the effects of RMRP on the aggressive phenotype of NSCLC. To this end, we overexpressed and knocked down the expression of RMRP in A549 and H1299. The transfection efficiency was confirmed by qRT-PCR (Figure 2A). Functionally, CCK8 assay and colony formation assay revealed that knockdown of RMRP inhibited the proliferation of A549 and H1299 (Figure 2B and C). The detection of PCNA, a marker of cell proliferation, also showed that knockdown of RMRP negatively regulated expression of PCNA (P<0.01; Figure 2D). Furthermore, the results of the Transwell chamber assay showed that the knockdown of RMRP weakened the invasion (Figure 2E) and migration (Figure 2F) ability of A549 and H1299 cells, while the overexpression of RMRP showed the opposite phenomenon. To further evaluate the metastatic potential, we measured the expression levels of a variety of crucial proteins. As a result, the RMRP knockdown negatively regulated the protein expression of Cox-2, MMP2, and MMP9, while overexpression of RMRP led to up-regulation (Figure 2G). Overall, our findings demonstrated that RMRP promotes NSCLC cell proliferation and metastasis potential.
RMRP Promotes NSCLC Cell Growth in vivo
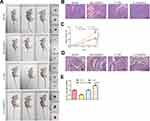
We further explored the effects of RMRP on the proliferation and metastatic potential in the NSCLC mice model. To study the effects of RMRP on the tumorigenicity of NSCLC mice models, the overexpressed- and silenced-A549 cells were subcutaneously injected into the right side of Balb/c nude mice. After four weeks, the tumor volume was observed. As revealed in Figure 3A and C, the tumors of the overexpression group were bigger than that of the control group (P<0.01), whereas the knockdown group observed the opposite trend (P<0.01). As shown in H&E staining of tumor tissues, the density of tumor cells of the RMRP overexpressed group was higher compared to the control group (Figure 3B). Moreover, the results of lung tissue H&E staining showed that vast area of cancer cell metastasis was observed in the overexpression group, while almost no metastasis was observed in the knockdown of the RMRP group (Figure 3D). As expected, the expression level of RMRP in tumor cells in vivo was consistent with that after transfection in vitro (Figure 3E). These results suggested that lncRNA RMRP promotes NSCLC cell proliferation, migration, and invasion in vivo.
RMRP Targets on the miR-613/NFAT5 Axis in the NSCLC Progression
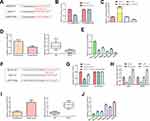
To further explore the molecular mechanism of RMRP promoting the migration and proliferation of NSCLC cells, we predicted that miR-613 was the target of lncRNA RMRP using the biological target prediction online tool TargetScan (Figure 4A). To verify this prediction, psiCHECK-2-RMRP-WT and psiCHECK-2-RMRP-Mut vectors were transfected into A549 cells, and the luciferase activity was determined after adding miR-613 mimic or mimic control for 48 hours. The luciferase activity of the RMRP-WT construct was decreased under the overexpression of miR-613, but its mutant construct was not altered (Figure 4B). Furthermore, we proved that the overexpression of RMRP inhibited the expression of miR-613 (Figure 4C). Moreover, the qRT-PCR analysis was conducted to detect the levels of miR-613 in 80 matched lung cancer tissues and corresponding paracancerous tissues. The expression of miR-613 in tumor tissues was lower than that in paracancerous tissues, which in advanced cases (stage III and IV) was lower than that in early-stage cases (stage I and II) (Figure 4D). These results indicated that the abnormal expression of miR-613 might be related to the progression of cancer. Besides, the levels of miR-613 in HCC827, 16HBE, H1299, H1975, and A549 cells were significantly decreased, compared with HCT116 cells (P<0.01; Figure 4E). These findings demonstrated that RMRP targets miR-613 and down-regulated the expression of miR-613 in NSCLC cells.
Next, we predicted that NFAT5 was the target of miR-613 using TargetScan (Figure 4F). Because of the potential roles of NFAT5 in NSCLC,16 we further verified this prediction. Then, WT and mutant 3ʹUTR region of NFAT5 vectors were transfected into RMRP overexpressed A549 cells, and the luciferase activity was determined by transfected with miR-613. The luciferase activity of the psiCHECK-2-NFAT5-WT construct was decreased under the miR-613 overexpression, whereas its mutant construct stayed unchanged (Figure 4G). Moreover, the luciferase activity of NFAT5 in A549 with miR-613 and RMRP overexpression was significantly increased (P<0.01; Figure 4G), compared to the only miR-613 overexpressed group, indicating that RMRP partly blocked the miR-613-induced down-regulation of NFAT5. Besides, the qRT-PCR analysis revealed that the RMRP overexpression up-regulated the mRNA level of NFAT5 in A549 and H1299 cells (Figure 4H). Additionally, the expression of NFAT5 in tumor tissues was significantly higher than that in paracancerous tissues, which in advanced cases (stage III and IV) was higher than that in early-stage cases (stage I and II) (Figure 4I). These results indicated that the over-expression of NFAT5 might be related to the progression of cancer. As expected, the mRNA levels of NFAT5 in HCC827, 16HBE, H1299, H1975, and A549 cells were significantly up-regulated, compared with HCT116 cells (P<0.01; Figure 4J). These results demonstrated that RMRP up-regulated the mRNA level of NFAT5 by sponging the miR-613 in NSCLC cells.
Inhibition of miR-613/NFAT5 Regulatory Axis Reverses the Role of RMRP Silencing-Induced on NSCLC Cells
Next, we investigated the effects of the miR-613/NFAT5 regulatory axis on the RMRP-silenced NSCLC cells. The results revealed that the RMRP knockdown inhibited the proliferation of A549 and H1299 cells (P<0.01; Figure 5A), whereas miR-613 inhibitor significantly neutralized RMRP silencing-inhibited cell proliferation (P<0.01). Moreover, the cell viability of the sh-RMRP/miRNA-613 inhibitor/sh-NFAT5 group was higher than that of the sh-RMRP/miRNA-613 inhibitor group (P<0.01). The trend of subsequent colony formation assay confirmed these results (P<0.01; Figure 5B). Furthermore, the detection of PCNA showed that miR-613 inhibition increased PCNA expression, which was reduced by RMRP silencing (P<0.01; Figure 5C). Besides, the PCNA expression of the sh-RMRP/miRNA-613 inhibitor/sh-NFAT5 group was significantly lower than that of the sh-RMRP/miRNA-613 inhibitor group (P<0.01). Also, we studied the effects of the miR-613/NFAT5 regulatory axis on the migration and invasion of RMRP-silenced NSCLC cells. As demonstrated in Figure 5D and E, miR-613 inhibition promoted the metastasis potential of RMRP-silenced A549 and H1299 cells (P<0.01), whereas NFAT5 knockdown significantly reversed this trend (P<0.01). At the molecular level, miR-613 inhibition reversed the down-regulation of Cox-2, MMP2, and MMP9 caused by RMRP silencing, while NFAT5 knockdown partially blocked the miR-613 inhibition (P<0.01; Figure 5F). The above data suggested that the inhibition of the miR-613/NFAT5 regulatory axis could reverse the effects of RMRP silencing on NSCLC progression, indicating that the miR-613/NFAT5 axis was direct downstream of RMRP.
Discussion
Through the location of large numbers of coding and non-coding genes, it has been proved that these genes play essential roles in the occurrence and tumorigenicity of NSCLC.17,18 However, the pathophysiological mechanism of the development of NSCLC is not clearly elucidated. In recent years, the research literature has highlighted the importance of lncRNAs in cancer biology.19,20 Although the research on lncRNAs has been increasing, only part of the lncRNAs function was clarified.
RMRP, which was initially identified in mouse, can process RNA transcripts complementary to light chains to produce RNA primers for heavy strand DNA replication.21 Moreover, RMRP is a critical factor for dysfunctional mutations in cartilage-hair dysplasia.22 In addition to these established functions, RMRP recently is significantly up-regulated in several tumor cells, such as colon cancer,23 gastric cancer,24 breast cancer,25 etc. Nevertheless, the function of RMRP in the development of NSCLC is poorly understood. In this report, our findings suggested that the level of RMRP in lung cancer and NSCLC cell lines was both higher than that in healthy tissues. Besides, the RMRP overexpression was positively corresponded with the clinical stage and overall survival in NSCLC patients, suggesting that RMRP might be a potential early diagnostic marker of NSCLC.
In further investigation, we found that the cell viability of NSCLC cells H1299 and A549 increased with the overexpression of RMRP, whereas the knockdown of RMRP showed the opposite trend. Consistent with the decrease of cell viability, the protein level of PCNA in RMRP silenced NSCLC cells decreased by about 60%. Similarly, consistent with the increase of cell viability, the protein level of PCNA in RMRP overexpressed NSCLC cells increased by about 40%. These data indicate that the proliferation of NSCLC cells was positively regulated by RMRP. Furthermore, we studied the effects of RMRP on the metastasis potential of NSCLC cells in vitro. The results showed that RMRP silencing significantly inhibited the metastasis potential of NSCLC cells. To further investigate the effect of RMRP on the tumorigenesis of NSCLC cells in vivo, we injected RMRP overexpressed- and silenced-A549 into the right side of BALB/c nude mice respectively. Obvious tumorigenesis was observed within three days, and the tumor volume was measured once a week. Consistent with in vitro experiments, the growth and migration of RMRP silenced NSCLC cells in nude mice significantly decreased. Based on the above data, we found that no matter in vivo or in vitro lncRNA knockdown of RMRP inhibits proliferation and metastasis potential of NSCLC cells.
miR-613 was initially reported to be related to lipid metabolism in HepG2 cells26 and macrophages.27 In recent years, lots of studies suggested that miR-613 may be associated with tumorigenesis.28–30 Zhu et al. reported that MiR-613 directly suppressed CXCR4 expression and then decreased the proliferation, migration, and induced apoptosis of osteosarcoma cells.31 Another report found that lncRNA CYTOR promoted the invasion and migration of nasopharyngeal carcinoma cells by targeting the miR-613/ANXA2 axis.32 Of note, the target gene of miR-613 in lung cancer was recently reported. Li et al. reported that miR-613 induced cell cycle arrest in NSCLC cells by sponging CDK4.14 In this report, it was found that RMRP negatively regulated the level of miR-613, which was related to its interaction with miR-613. The mutation of miR-613 3ʹ-UTR weakened the RMRP mediated inhibition of miR-613 expression. The above results indicated that miR-613 is the functional target of RMRP in NSCLC cells. More importantly, our data revealed that the nuclear factor of activated T cell 5 (NFAT5) is a functional target of miR-613.
NFAT5 is found initially as a transcription factor of IL-2 in T cells and plays a substantial role in activating gene transcription in immune response.33 In recent years, due to its extensive functions in cell growth and differentiation, investigators have broadened its role in tumorigenesis and development.34 The deletion of NFAT5 subtypes arrests the cell cycle and inhibits the progression of melanoma cells.35 Guo et al. find that the level of NFAT5 was up-regulated in lung adenocarcinoma cells, and NFAT5 promotes the proliferation and migration of cancer cells by up-regulating AQP5.36 More evidence had shown that the function of NFAT5 could be regulated by miRNAs. Meng et al demonstrated that the miR-194 down-regulated the level of NFAT5, which inhibited the progression of lung cancer cells.16 Therefore, NFAT5 may be profoundly involved in the development of lung carcinoma, which is worthy of further exploration. In this study, the data suggested that the inhibition of the miR-613/NFAT5 regulatory axis reversed the effects of RMRP silencing on NSCLC cells, indicating that the miR-613/NFAT5 axis was direct downstream of RMRP.
In conclusion, RMRP promotes NSCLC cell proliferation and metastasis by targeting the miR-613/NFAT5 axis. Moreover, knockdown of RMRP inhibited the metastasis potential of A549 and H1299 cells, which was reversed by inhibition of the miR-613/NFAT5 regulatory axis. In short, lncRNA RMRP promotes cell proliferation and invasion through the miR-613/NFAT5 axis in NSCLC cells.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi:10.3322/caac.21208
2. Lee DH. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): the road to a success, paved with failures. Pharmacol Therapeut. 2017;174:1–21. doi:10.1016/j.pharmthera.2017.02.001
3. Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52(Pt 1):103–109. doi:10.1016/j.semcancer.2017.11.019
4. Kubo H, Suzuki T, Matsushima T, et al. Cyclin-dependent kinase-specific activity predicts the prognosis of stage I and stage II non-small cell lung cancer. BMC Cancer. 2014;14(1):755. doi:10.1186/1471-2407-14-755
5. Lau E. Zooming in on lncRNA functions. Nat Rev Genet. 2014;15(9):574–575. doi:10.1038/nrg3795
6. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
7. Rosenbluh J, Nijhawan D, Chen Z, et al. RMRP is a non-coding RNA essential for early murine development. PLoS One. 2011;6(10):e26270. doi:10.1371/journal.pone.0026270
8. Zhou N, He Z, Tang H, Jiang B, Cheng W. LncRNA RMRP/miR-613 axis is associated with poor prognosis and enhances the tumorigenesis of hepatocellular carcinoma by impacting oncogenic phenotypes. Am J Transl Res. 2019;11(5):2801.
9. Cao H, Liu Z, Huang P, Yue Y, Xi J. lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206. Eur Rev Med Pharmaco. 2019;23(3):1012.
10. Wang J, Xiao T, Zhao M. MicroRNA-675 directly targets MAPK1 to suppress the oncogenicity of papillary thyroid cancer and is sponged by long non-coding RNA RMRP. Oncotargets Ther. 2019;12:7307–7321. doi:10.2147/OTT.S213371
11. Wang Y, Luo X, Liu Y, Han G, Sun D. Long noncoding RNA RMRP promotes proliferation and invasion via targeting miR‐1‐3p in non–small‐cell lung cancer. J Cell Biochem. 2019;120(9):15170–15181. doi:10.1002/jcb.28779
12. Mei J, Xu R, Hao L, Zhang Y. MicroRNA-613: a novel tumor suppressor in human cancers. Biomed Pharmacother. 2020;123:109799. doi:10.1016/j.biopha.2019.109799
13. Chen X, Xu Y, Liao X, et al. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol. 2016;37(9):11927–11936. doi:10.1007/s13277-016-5052-8
14. Li D, Li D, Liu D, Tang X. MiR-613 induces cell cycle arrest by targeting CDK4 in non-small cell lung cancer. Cell Oncol. 2016;39(2):139–147. doi:10.1007/s13402-015-0262-4
15. National Research Council (US) Institute for Laboratory Animal Research. Guide for the Care and Use of Laboratory Animals. Washington (DC): National Academies Press (US); 1996.
16. Meng X, Li Z, Zhou S, Xiao S, Yu P. miR-194 suppresses high glucose-induced non-small cell lung cancer cell progression by targeting NFAT5. Thorac Cancer. 2019;10(5):1051–1059. doi:10.1111/1759-7714.13038
17. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45(8):1895–1910. doi:10.1016/j.biocel.2013.05.030
18. Moreno-Moya JM, Vilella F, Simón C. MicroRNA: key gene expression regulators. Fertil Steril. 2014;101(6):1516–1523. doi:10.1016/j.fertnstert.2013.10.042
19. Xue Y, Ma G, Zhang Z, et al. A novel antisense long noncoding RNA regulates the expression of MDC1 in bladder cancer. Oncotarget. 2015;6(1):484–493. doi:10.18632/oncotarget.2861
20. Ma M, Chu B, Zhang Y, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6(1):e1583–e1583. doi:10.1038/cddis.2014.541
21. Chang DD, Clayton DA. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56(1):131–139. doi:10.1016/0092-8674(89)90991-4
22. Martin AN, Li Y. RNase MRP RNA and human genetic diseases. Cell Res. 2007;17(3):219–226. doi:10.1038/sj.cr.7310120
23. Park J, Jeong S. Wnt activated beta-catenin and YAP proteins enhance the expression of non-coding RNA component of RNase MRP in colon cancer cells. Oncotarget. 2015;6(33):34658–34668. doi:10.18632/oncotarget.5778
24. Shao Y, Ye M, Li Q, et al. LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7(25):37812–37824. doi:10.18632/oncotarget.9336
25. Rheinbay E, Parasuraman P, Grimsby J, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547(7661):55–60. doi:10.1038/nature22992
26. Zhong D, Zhang Y, Zeng Y, et al. MicroRNA-613 represses lipogenesis in HepG2 cells by downregulating LXRα. Lipids Health Dis. 2013;12(1):32. doi:10.1186/1476-511X-12-32
27. Zhao R, Feng J, He G. miR-613 regulates cholesterol efflux by targeting LXRα and ABCA1 in PPARγ activated THP-1 macrophages. Biochem Bioph Res Co. 2014;448(3):329–334. doi:10.1016/j.bbrc.2014.04.052
28. Chandrakesan P, Weygant N, May R, et al. Dclk1 facilitates intestinal tumor growth via enhancing pluripotency and epithelial mesenchymal transition. Oncotarget. 2014;5(19):9269–9280. doi:10.18632/oncotarget.2393
29. Sureban SM, May R, Weygant N, et al. XMD8-92 inhibits pancreatic tumor xenograft growth via a DCLK1-dependent mechanism. Cancer Lett. 2014;351(1):151–161. doi:10.1016/j.canlet.2014.05.011
30. Sun B, Ke K, Liu D, et al. Long noncoding RNA SNHG14 acts as an oncogene in prostate cancer via targeting miR-613. Eur Rev Med Pharmaco. 2020;24(2):633.
31. Zhu Y, Tang L, Zhao S, et al. CXCR4-mediated osteosarcoma growth and pulmonary metastasis is suppressed by MicroRNA-613. Cancer Sci. 2018;109(8):2412–2422. doi:10.1111/cas.13653
32. Chen W, Du M, Hu X, et al. Long noncoding RNA cytoskeleton regulator RNA promotes cell invasion and metastasis by titrating miR‐613 to regulate ANXA2 in nasopharyngeal carcinoma. Cancer Med-Us. 2019;9(3):1209–1219. doi:10.1002/cam4.2778
33. Shaw J, Utz P, Durand D, et al. Identification of a putative regulator of early T cell activation genes. Science. 1988;241(4862):202–205. doi:10.1126/science.3260404
34. Shou J, Jing J, Xie J, et al. Nuclear factor of activated T cells in cancer development and treatment. Cancer Lett. 2015;361(2):174–184. doi:10.1016/j.canlet.2015.03.005
35. Kim D, Kim K, Ramakrishna S. NFAT5 promotes in vivo development of murine melanoma metastasis. Biochem Bioph Res Co. 2018;505(3):748–754. doi:10.1016/j.bbrc.2018.09.171
36. Guo K, Jin F. NFAT5 promotes proliferation and migration of lung adenocarcinoma cells in part through regulating AQP5 expression. Biochem Bioph Res Co. 2015;465(3):644–649. doi:10.1016/j.bbrc.2015.08.078
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.