Back to Journals » OncoTargets and Therapy » Volume 13
lncRNA RHPN1-AS1 Promotes Ovarian Cancer Growth and Invasiveness Through Inhibiting miR-1299
Authors Zhao L, Liu T, Zhang X, Zuo D, Liu C
Received 2 February 2020
Accepted for publication 1 May 2020
Published 10 June 2020 Volume 2020:13 Pages 5337—5344
DOI https://doi.org/10.2147/OTT.S248050
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Lin Zhao, Ting Liu, Xingna Zhang, Donghua Zuo, Chunna Liu
Department of Gynaecology, Linyi Cancer Hospital, Linyi 276000, People’s Republic of China
Correspondence: Chunna Liu
Department of Gynaecology, Linyi Cancer Hospital, Linyi 276000, People’s Republic of China
Email [email protected]
Background: Ovarian cancer (OC) is a big threat for public health. However, the molecular mechanism underlying OC development and progression remains unclear. Although the importance of lncRNA in cancer has been proven, how lncRNA is involved in OC is waiting for further investigation.
Materials and Methods: qRT-PCR was performed to test expression level. CCK8 and colony formation were conducted to analyze proliferation. Transwell was conducted to measure migration and invasion. Luciferase reporter assay and pulldown assay were utilized to validate RNA interaction.
Results: lncRNA RHPN1-AS1 was highly expressed in OC tissues. RHPN1-AS1 was positively correlated with OC progression and its high expression indicated a low survival rate. Moreover, knockdown of RHPN1-AS1 significantly inhibited the proliferation, migration and invasion of OC cells, and bioinformatics analysis identified that miR-1299 was sponged by RHPN1-AS1 in OC cells. Knockdown of RHPN1-AS1 markedly promoted miR-1299 expression. Of note, inhibition of miR-1299 reversed the roles of RHPN1-AS1 silencing on suppressing proliferation, migration and invasion.
Conclusion: Our study demonstrates that RHPN1-AS1 promotes OC progression via sponging miR-1299, suggesting RHPN1-AS1 may be a novel therapeutic target.
Keywords: ovarian cancer, RHPN1-AS1, miR-1299, proliferation, invasion
Introduction
Ovarian cancer (OC) is a very deadly gynaecological tumor.1 Patients are usually diagnosed with OC at the advanced stage.2 And high rates of recurrence and metastasis further contribute to the mortality of OC patients.3 At present, the main strategies for OC treatment include surgery, chemotherapy and radiotherapy.4 However, the molecular mechanism underlying OC tumorigenesis remains unclear and the outcomes of OC patients are very poor.5 The five-year survival rate of OC patients remains lower than 35%.6 Thus, determining the factors regulating OC development and identifying effective therapeutic targets are urgently required.
Long noncoding RNAs (lncRNAs) are characterized with no protein-coding potential and have over 200 nucleotides in length.7 lncRNAs are tissue-specific expressed and exert vital functions in various biological processes.8 lncRNAs could regulate development, immune response and tumorigenesis via interacting with several molecules, such microRNAs and proteins.9 In cancer, several lncRNAs are found to be aberrantly expressed and correlated with tumor progression. For example, lncRNA GAS5 is downregulated in OC and serve as a tumor suppressor to inhibit cancer development.10 lncRNA MACC1-AS1 is upregulated in pancreatic cancer and promotes tumor growth via activating PAX8-NOTCH1 signal.11 Additionally, lncRNA LINC00668 overexpression contributes to breast cancer development via regulating apoptosis and proliferation.12 RHPN1-AS1 is a poorly researched lncRNA. A report has showed that RHPN1-AS1 promotes melanoma development.13 Furthermore, RHPN1-AS1 also regulates the gefitinib resistance of lung cancer cells and promotes breast cancer metastasis.14,15 Nevertheless, the roles of RHPN1-AS1 in OC are largely unknown.
In this study, we aimed to elucidate the physiological function of RHPN1-AS1 in OC progression. We found that RHPN1-AS1 expression was elevated in OC tissues and cell lines. Moreover, upregulation of RHPN1-AS1 was associated with lymph node metastasis and poor prognosis. Knockdown of RHPN1-AS1 inhibited OC progression via inhibiting proliferation, migration and invasion. We also showed that RHPN1-AS1 interacted with miR-1299 to inhibit its level. And we demonstrated that miR-1299 is a tumor suppressor and acted at the downstream of RHPN1-AS1 in OC. Taken together, our research reveals the novel functions of RHPN1-AS1 in OC and illustrated its molecular mechanism.
Materials and Methods
Tissue Samples
Fifty seven OC tissues and their adjacent normal controls were obtained from Linyi Cancer Hospital. Patients’ tissues that were treated with radiotherapy or chemotherapy prior to surgery were excluded. Other samples were included. All tissues were stored in liquid nitrogen. Association between RHPN1-AS1 Expression and clinical features in ovarian cancer tissues was analyzed in Table 1. This study was approved by the Ethics Committee of Linyi Cancer Hospital (No. 201,801,240,241). Written informed consent was obtained from each patient.
 |
Table 1 Association Between RHPN1-AS1 Expression and Clinical Features in Ovarian Cancer Patients (n=57) |
Cell Lines and Transfection
The human OC cell lines (OVCAR5, OVCAR3, A2780 and SKOV3 cells) and normal ovarian epithelial cell line IOSE80 were purchased from American Type Culture Collection (ATCC, USA). These cells were cultured using RPMI-1640 medium (Gibco, Grand Island, NY, USA), supplemented with 10% FBS (HyClone, Logan, UT, USA) in a humidified atmosphere, at 37 °C with 5% CO2.
Specific siRNA RHPN1-AS1 and scrambled siRNA control (si-NC) were purchased from GenePharma (Shanghai, China). miR-1299 mimics, miR-1299 inhibitors and negative controls were from RiboBio (Guangzhou, China). All oligonucleotides were transfected into cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
qRT-PCR
RNA isolation was performed using Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA concentration was determined using the Nanodrop spectrophotometer (ND-100; Thermo Scientific, Waltham, MA). SYBR Premix Ex Taq and TaqMan gene expression assays (Applied Biosystems, Foster City, CA, USA) were utilized to analyze RHPN1-AS1 and GAPDH expression. TaqMan MicroRNA Reverse Transcription kit and Taqman Universal Master Mix II (Applied Biosystems) were used for miR-1299 and U6 analyses. Relative expression was calculated according to the 2−ΔΔCt method. The primer sequences were as follows: RHPN1-AS1 (Forward, 5ʹ-CTAGCCAGGAGGTTTCGC-3ʹ and reverse, 5ʹ-TCCGCAACAAGCACACA-3ʹ) and GAPDH (Forward, 5ʹ-CACCCACTCCTCCACCTTTG-3ʹ and reverse, 5ʹ-CCACCACCCTGTTGCTGTAG-3ʹ).
CCK8 Assay
Cell Counting Kit-8 (CCK8; Roche, Basel, Switzerland) was used for analyzing cell proliferation. Cells were seeded into the 96-well plates and cultured for several days. Then 10 μL CCK8 solution was added and incubated for 2 h. The absorbance was then measured at 450 nm.
Colony Formation Assay
500 cells per well was plated in the 6-well plates and cultured for two weeks. Then cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Colony (over 50 cells) number was then calculated.
Transwell Assay
Cell migration and invasion was tested using Transwell assay as reported before.16 Cells were seeded into the upper chamber (pore size: 8 μm, BD Biosciences, USA) with 200 μL serum-free medium. The lower chamber was filled with 600 μL complete culture medium. After cultured for 24 h, the cells migrated or invaded into the lower chamber were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Cell numbers were quantified using a light microscope. Matrigel was used only for invasion assay.
RNA Pulldown
RNA pulldown was performed using biotin-labeled miR-1299-WT or miR-1299-Mut as previously reported.17 In brief, biotin-labeled miR-1299-WT, miR-1299-Mut or biotin-NC was incubated with tumor cell lysates, followed by incubation with Streptavidin-coupled beads. Precipitated RNAs were purified and analyzed by qRT-PCR.
RNA Immunoprecipitation (RIP)
RIP assay was completed as described before.17 In brief, tumor cell lysates were incubated with anti-Ago2 or IgG, followed by incubation with Protein A/G beads. Then precipitated RNAs were purified and analyzed through qRT-PCR.
Luciferase Activity Assay
The RHPN1-AS1 sequence containing putative miR-1299 binding site was inserted into pmirGLO vector (Promega, Madison, WI, USA). Then the RHPN1-AS1-WT or RHPN1-AS1-Mut reporter and miR-1299 mimics were co-transfected into OC cells. 48 h later, the relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocols. Renilla luciferase activity was the transfection control.
Statistical Analysis
Statistical analysis was carried out with the statistical program SPSS 13.0 (IBM Corp, Armonk, NY). Results were displayed as mean ± standard deviation and analyzed using the t-test and one-way analysis of variance (ANOVA). P value <0.05 was considered statistically significant.
Results
lncRNA RHPN1-AS1 Was Upregulated in OC Tissues and Cell Lines
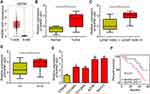
We analyzed the expression of RHPN1-AS1 in OC tissues according to the GEPIA database. RHPN1-AS1 expression was upregulated in OC tissues compared to normal tissues (Figure 1A). qRT-PCR also confirmed that RHPN1-AS1 levels were increased in 57 OC tissues compared to corresponding normal controls (Figure 1B). Interestingly, we found that RHPN1-AS1 expression was positively correlated with lymph node metastasis and clinical stage (Figure 1C and D). Similarly, qRT-PCR showed that RHPN1-AS1 level was elevated in OC cell lines (Figure 1E). Then we divided these samples into two subgroups based on RHPN1-AS1 expression (the median value of RHPN1-AS1 as the cut-off). We found that RHPN1-AS1 overexpression OC patients displayed a rather low survival rate (Figure 1F), indicating RHPN1-AS1 might be a prognostic biomarker.
Silencing of RHPN1-AS1 Inhibited OC Proliferation, Migration and Invasion
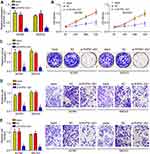
To analyze the function of RHPN1-AS1, we chose A2780 and SKOV3 cells to perform experiments. Firstly, we verified the knockdown efficiency of RHPN1-AS1 in A2780 and SKOV3 cells (Figure 2A). Then CCK8 assay was performed. We found that RHPN1-AS1 knockdown significantly inhibited the proliferation of A2780 and SKOV3 cells (Figure 2B). Similarly, decreased expression of RHPN1-AS1 led to reduced colony numbers (Figure 2C). Moreover, RHPN1-AS1 knockdown also attenuated the migration and invasion of A2780 and SKOV3 cells (Figure 2D and E). Hence, RHPN1-AS1 promotes OC proliferation, migration and invasion.
RHPN1-AS1 Sponged miR-1299 in OC
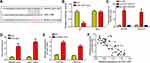
To explore the mechanism, we utilized bioinformatics analysis (miRDB and LncBase v.2) to search the potential targets of RHPN1-AS1. We identified miR-1299 because it has the highest score (Figure 3A). Then luciferase reporter assay was performed with A2780 cells. The result showed that the activity of RHPN1-AS1-WT was inhibited by miR-1299 mimics (Figure 3B). RNA pulldown assay indicated that RHPN1-AS1 was precipitated by bio-miR-1299-WT but not miR-1299-Mut or bio-NC (Figure 3C). RIP assay demonstrated that RHPN1-AS1 and miR-1299 were enriched by anti-Ago2 (Figure 3D). Above data suggested that RHPN1-AS1 directly interacted with miR-1299. We then found that RHPN1-AS1 silencing led to miR-1299 upregulation in OC cells (Figure 3E). And RHPN1-AS1 level was negatively correlated with miR-1299 in OC tissues (Figure 3F). In conclusion, RHPN1-AS1 sponged miR-1299 in OC.
RHPN1-AS1 Promoted OC Development via Repressing miR-1299
Afterwards, we sought to determine whether RHPN1-AS1 regulates OC progression by miR-1299. We found that miR-1299 was downregulated in OC tissues (Figure 4A). And miR-1299 expression was downregulated after transfection with miR-1299 inhibitors (Figure 4B). We then performed rescue assay. We found that miR-1299 inhibitors rescued the proliferation, migration and invasion of OC cells transfected with RHPN1-AS1 siRNA (Figure 4C–F). Furthermore, miR-1299 inhibitors alone could promote proliferation, migration and invasion of OC cells (Figure 4C–F). Thus, miR-1299 exerts an opposite roles as RHPN1-AS1 in OC.
Discussion
Accumulating studies have demonstrated that lncRNA play vital functions in tumor initiation and progression and may potential novel therapeutic targets for tumor intervention.18,19 In our research, we provided new data, demonstrating that RHPN1-AS1 is highly expressed in OC tissues and promotes OC progression via sponging miR-1299.
At first, we searched the GEPIA database to investigate the expression pattern of RHPN1-AS1 in OC tissues. We found that RHPN1-AS1 was highly expressed in OC tissues compared to normal tissues. To confirm it, we collected 57 pairs of OC tissues and normal controls. Through qRT-PCR, we verified that RHPN1-AS1 was upregulated in both OC tissues and cell lines. In the researched four OC cell lines, RHPN1-AS1 was upregulated in all OC cell lines. Moreover, RHPN1-AS1 expression was positively correlated with metastasis and clinical stage. And RHPN1-AS1 upregulation is linked with a rather low survival rate in OC patients. All these findings suggest RHPN1-AS1 may be an important regulator in OC progression. In previous work, RHPN1-AS1 was found to promote melanoma development.13 The authors found that RHPN1-AS1 was upregulated in melanoma tissues.13 Besides, RHPN1-AS1 is also identified to be upregulated in lung cancer and breast cancer.14,15 By combining with previous works, RHPN1-AS1 may be overexpressed in various human cancer tissues but not one. And it may be a critical tumor-associated lncRNA.
Afterwards, we used siRNA to knock down RHPN1-AS1 and explore its physiological function in OC. We selected A2780 and SKOV3 cells and successfully silenced RHPN1-AS1 in these two cell lines. By using biochemical assays, we demonstrated that RHPN1-AS1 knockdown displayed an important anti-cancer role by regulating OC proliferation, migration and invasion. This finding is not surprising because RHPN1-AS1 was also found to regulate tumor proliferation and metastasis in several other cancers, such as melanoma and breast cancer.13,15 Nevertheless, our present research demonstrated the roles of RHPN1-AS1 in OC for the first time.
Another important finding of our work may be the identification of the endogenous competing mechanism between RHPN1-AS1 and miR-1299. We used two online tools to predict their interaction. Then a series of experiments were performed to confirm their direct interaction. We performed luciferase reporter assay, RNA pulldown assay and RIP assay. All these assays indicated that RHPN1-AS1 bound to miR-1299 directly. Moreover, we found that RHPN1-AS1 knockdown caused upregulation of RHPN1-AS1 in OC cells. And their expression was negatively correlated in OC tissues. miR-1299 is a rarely known miRNA. A previous study showed that miR-1299 inhibits hepatocellular carcinoma growth via inactivating CDK6.20 miR-1299 also suppresses colon cancer development.21 Additionally, miR-1299 also exerts anti-cancer roles in prostate cancer, cholangiocarcinoma and breast cancer.22–24 However, the role of miR-1299 in OC has not been determined. Thus, we further explored how RHPN1-AS1/miR-1299 axis regulates OC progression. We found that miR-1299 was downregulated in OC tissues. And miR-1299 inhibitors rescued the proliferation, migration and invasion of OC cells with RHPN1-AS1 siRNA transfection. More importantly, miR-1299 inhibition alone promoted OC cell proliferation, migration and invasion. Thus, miR-1299 is a critical suppressor of OC progression.
In conclusion, our work demonstrated that RHPN1-AS1 promotes OC progression via targeting miR-1299. And RHPN1-AS1 is a prognostic biomarker for OC patients. However, there are some limitations in our study. For example, in vivo assay is required to validate the roles of RHPN1-AS1 in the future.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yokoi A, Matsuzaki J, Yamamoto Y, et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9:4319.
2. Pisanic TR
3. Shah HK, Bhat MA, Sharma T, Banerjee BD, Guleria K. Delineating potential transcriptomic association with organochlorine pesticides in the etiology of epithelial ovarian cancer. Open Biochem J. 2018;12:16–28. doi:10.2174/1874091X01812010016
4. Yang M, Zhai Z, Guo S, Li X, Zhu Y, Wang Y. Long non-coding RNA FLJ33360 participates in ovarian cancer progression by sponging miR-30b-3p. Onco Targets Ther. 2019;12:4469–4480. doi:10.2147/OTT.S205622
5. Oza AM, Cibula D, Benzaquen AO, et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised Phase 2 trial. Lancet Oncol. 2015;16:87–97. doi:10.1016/S1470-2045(14)71135-0
6. Heindl A, Khan AM, Rodrigues DN, et al. Microenvironmental niche divergence shapes BRCA1-dysregulated ovarian cancer morphological plasticity. Nat Commun. 2018;9:3917. doi:10.1038/s41467-018-06130-3
7. Leisegang MS. LET’s sponge: how the lncRNA PFL promotes cardiac fibrosis. Theranostics. 2018;8:874–877. doi:10.7150/thno.23364
8. Krause HM. New and prospective roles for lncRNAs in organelle formation and function. Trends Genet. 2018;34:736–745. doi:10.1016/j.tig.2018.06.005
9. Liu B, Ye B, Yang L, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499–508. doi:10.1038/ni.3712
10. Long X, Song K, Hu H, et al. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res. 2019;38:345. doi:10.1186/s13046-019-1329-2
11. Qi C, Xiaofeng C, Dongen L, et al. Long non-coding RNA MACC1-AS1 promoted pancreatic carcinoma progression through activation of PAX8/NOTCH1 signaling pathway. J Exp Clin Cancer Res. 2019;38:344. doi:10.1186/s13046-019-1332-7
12. Qiu X, Dong J, Zhao Z, Li J, Cai X. LncRNA LINC00668 promotes the progression of breast cancer by inhibiting apoptosis and accelerating cell cycle. Onco Targets Ther. 2019;12:5615–5625. doi:10.2147/OTT.S188933
13. Lu L, Yu X, Zhang L, et al. The long non-coding RNA RHPN1-AS1 promotes uveal melanoma progression. Int J Mol Sci. 2017;18. doi:10.3390/ijms18010226
14. Li X, Zhang X, Yang C, Cui S, Shen Q, Xu S. The lncRNA RHPN1-AS1 downregulation promotes gefitinib resistance by targeting miR-299-3p/TNFSF12 pathway in NSCLC. Cell Cycle. 2018;17:1772–1783. doi:10.1080/15384101.2018.1496745
15. Zheng S, Lv P, Su J, Miao K, Xu H, Li M. Silencing of the long non-coding RNA RHPN1-AS1 suppresses the epithelial-to-mesenchymal transition and inhibits breast cancer progression. Am J Transl Res. 2019;11:3505–3517.
16. Chen J, Lin Y, Jia Y, Xu T, Wu F, Jin Y. LncRNA HAND2-AS1 exerts anti-oncogenic effects on ovarian cancer via restoration of BCL2L11 as a sponge of microRNA-340-5p. J Cell Physiol. 2019;234(12):23421–23436. doi:10.1002/jcp.28911
17. Li W, Ma S, Bai X, Pan W, Ai L, Tan W. Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting as a competing endogenous RNA of microRNA-18a in ovarian cancer. J Cell Physiol. 2020;235(2):1141–1154.
18. Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1:93–109. doi:10.1016/j.trecan.2015.08.010
19. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi:10.1038/onc.2017.184
20. Zhu H, Wang G, Zhou X, et al. miR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed Pharmacother. 2016;83:792–797. doi:10.1016/j.biopha.2016.07.037
21. Wang Y, Lu Z, Wang NN, Zhang M, Zeng XD, Zhao W. MicroRNA-1299 is a negative regulator of STAT3 in colon cancer. Oncol Rep. 2017;37:3227–3234. doi:10.3892/or.2017.5605
22. Zhang FB, Du Y, Tian Y, Ji ZG, Yang PQ. MiR-1299 functions as a tumor suppressor to inhibit the proliferation and metastasis of prostate cancer by targeting NEK2. Eur Rev Med Pharmacol Sci. 2019;23:530–538. doi:10.26355/eurrev_201901_16865
23. Xu Y, Yao Y, Liu Y, et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging (Albany NY). 2019;11:1907–1917. doi:10.18632/aging.101872
24. Liu LH, Tian QQ, Liu J, Zhou Y, Yong H. Upregulation of hsa_circ_0136666 contributes to breast cancer progression by sponging miR-1299 and targeting CDK6. J Cell Biochem. 2019;120:12684–12693. doi:10.1002/jcb.28536
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.