Back to Journals » OncoTargets and Therapy » Volume 12
LncRNA PVT1 promotes proliferation, invasion and epithelial–mesenchymal transition of renal cell carcinoma cells through downregulation of miR-16-5p
Authors Ren Y, Huang W, Weng G, Cui P, Liang H, Li Y
Received 9 October 2018
Accepted for publication 7 February 2019
Published 5 April 2019 Volume 2019:12 Pages 2563—2575
DOI https://doi.org/10.2147/OTT.S190239
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yu Ren,1 Weiping Huang,2 Guobin Weng,1 Pinger Cui,1 Haote Liang,2 Yeping Li2
1Department of Urology, Ningbo Urology and Nephrology Hospital, Ningbo 315000, Zhejiang Province, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, Zhejiang Province, People’s Republic of China
Background: LncRNAs have recently emerged as vital regulators in the pathogenesis and development of various cancers. LncRNA PVT1 is reported to function as an oncogene in some tumors. However, the role of PVT1 in RCC remains unknown.
Purpose: To explore the potential effects of lncPVT1 on the development of renal cell carcinoma.
Methods: The expression of PVT1 in renal cancer cell lines and tissues was measured by qRT-PCR. The endogenous PVT1 was silenced by RNAi. Cell viabilities were measured by the MTT assay. The migration and invasion of cells were investigated by the transwell assay. The apoptosis of cells was measured by the Nucleosome ELISA and caspase-3 activity assays. The levels of proteins were measured by the western blot.
Results: We found that PVT1 was upregulated in RCC tissues compared with the adjacent normal tissues. PVT1 expression was closely correlated with TNM stage, Fuhrman grade, lymph node metastasis and tumor size. Kaplan–Meier analysis revealed that high expression of PVT1 was significantly associated with poor overall survival. In accordance, overexpression of PVT1 was observed in RCC cells comto HK-2 cell. Silencing of PVT1 significantly repressed cell viability, induced apoptosis and inhibited cell migration and invasion in vitro. Furthermore, bioinformatic analysis and luciferase reporter assay confirmed that miR-16-5p was a target of PVT1. Silencing of miR-16-5p mostly reversed the regulatory effects on RCC cells induced by downregulation of PVT1.
Conclusion: In summary, our study indicates that targeting PVT1 might represent a rational therapeutic strategy for RCC.
Keywords: renal carcinoma, PVT1, migration, invasion, apoptosis, miR-16-5p
Corrigendum for this paper has been published
Corrigendum for this paper has been published
Introduction
Renal cell carcinoma (RCC) is one of the most common cancers and accounts for about 3% of all human malignancies.1 The incidence of RCC has gradually increased over 2 decades.2 RCC is heterogeneous and can be divided into four subtypes as follows: clear cell RCC (ccRCC), chromophobe RCC (chRCC), renal oncocytoma (RO) and papillary RCC (pRCC). Among them, ccRCC is the most common subtype and accounts for around 70%–80% of all RCC cases.3 Despite the progress of novel therapeutic agents, it is still difficult to treat patients with metastatic RCC, and the prognosis remains poor.4 Therefore, a better understanding of the mechanisms involved in the development of RCC and more effective therapeutic strategies are urgently needed.
Long non-coding RNAs (lncRNAs) are a group of RNA molecules longer than 20 nucleotides that are not translated into proteins.5 Mounting evidence suggests that lncRNAs act as major regulators of various biological activities such as proliferation, homeostasis, differentiation, apoptosis and metastasis.6,7 LncRNAs could act as a competing endogenous RNA (ceRNA) and sponge micro-RNAs (miRNAs), thus affecting the expression of various proteins. Therefore, lncRNAs have been regarded as vital players in cancer research, and many studies have found that some lncRNAs function as tumor inhibitors, oncogenes, or both, under different circumstances.8 LncRNA plasmacytoma variant translocation 1 (PVT1) has been found to be dysregulated in various cancers. For example, PVT1 could promote tumor progression via interacting with FOXM1 in gastric cancer.9 Overexpression of PVT1 is a poor prognostic biomarker and promotes migration and invasion in colorectal cancer.10 PVT1 also promotes cervical cancer progression by acting as a molecular sponge to negatively regulate miR-424.11 However, little is known about the expression level and functional roles of PVT1 in RCC.
The aim of the present study is to investigate the role of PVT1 in RCC. We found that PVT is upregulated in RCC tissues and cells. We also examined the effects of PVT1 on cell proliferation, migration, invasion and apoptosis. Moreover, we found that PVT acted as a ceRNA by negatively regulating miR-16-5p. Taken together, our findings may contribute to the development of effective clinical interventions against RCC.
Materials and methods
Clinical samples
A total of 25 patients with pathologically confirmed clear cell RCC (ccRCC) were enrolled in this study. None of them received preoperative chemotherapy and/or radiotherapy. Tissues were frozen in liquid nitrogen immediately after surgery and subsequently stored at −80°C. They were classified according to the TNM classification and criteria of WHO. Written informed consent was obtained from all patients. The ethics committee of Wenzhou Medical University approved this study. The study was conducted in accordance with the Declaration of Helsinki.
Cell culture
Normal renal epithelial cells (HK-2) and renal cancer cell lines (A498, 786-O, ACHN and Caki-1) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were maintained in RPMI 1640 medium (HyClone, Logan, UT, USA) supplemented with 10% FBS, 100 U/mL of penicillin and 50 μg/mL of streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). All cells were maintained in a humidified incubator with 5% CO2 at 37°C.
RNA isolation and quantitative real-time (qRT-PCR)
Total RNA was isolated from tissues and cell lines using TRizol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The cDNA was synthesized from 200 ng of extracted total RNA using the First Strand cDNA Synthesis Kit (Takara, Dalian, China). For the measurement of miRNA expression, miRNA cDNA was synthesized using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). qRT-PCR was performed with SYBR Green (Takara), and the data collection was performed on the Applied Biosystem 7500 system. The primers were synthesized by Genescript (Nanjing, China). The relative expression level of the indicated genes was normalized to GAPDH or U6; expression fold changes were calculated using 2−ΔΔCt methods; and each reaction was performed in triplicate. Following primers are used for qRT-PCR: PVT1: forward 5′-ATAGATCCTGCCCTGTTTGC-3′; reverse 5′-CATTTCCTGCTGCCGTTTTC-3′; miR-16-5p: forward 5′-TCCACTCTAGCAGCACGTAAAT-3′; reverse 5′-TCACACTAAAGCAGCACAGTAAT-3′; and GAPDH: forward 5′-GGAGCGAGATCCCTCCAAAAT-3′; reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Plasmid construction, oligonucleotide and transfection
Two short hairpin RNA (shRNA) sequences targeting PVT1 (sh-PVT1 1 and sh-PVT1 2) or negative control (sh-NC) were designed by Genepharm Co. Ltd. (Shanghai, China). After annealing, double strands of shRNA were inserted into lentiviral pU6-Luc-Puro vector (Genepharm Co. Ltd.). pcDNA3.1 vectors containing full length PVT1 or mutant PVT1 were obtained from RioBio (Guangzhou, China) and transfected into cells using Lipofectamine 2000 (Thermo Fisher Scientific). The miR-16-5p mimics, inhibitor and negative control were synthesized by Genepharma (Shanghai, China) and transfected into cells using Lipofectamine 2000 (Thermo Fisher Scientific).
Dual luciferase reporter assay
The wild-type lncRNA-PVT1 and a mutant lncRNA-PVT1 sequence (mutant in miR-16-5p binding site) were cloned into pmirGLO plasmid. The pmirGLO-lncRNA-PVT1 or pmirGLO-lncRNA-PVT1-mut was co-transfected with miR-16-5p mimics or miRNA negative control (miR-NC) by Lipofectamine 2000 (Thermo Fisher Scientific). Luciferase activity was determined by the Dual-luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA). The luciferase activity was normalized to Renilla luciferase activity.
Cell viability assay
Cell viability was measured as described by Yu et al.12 Cells were seeded onto 96-well plates at a density of 5,000 per wells. After transfection, 20 μL of 5 mg/mL MTT (Sigma-Aldrich Co., St Louis, MO, USA) was added into each well and incubated for 4 h at 37°C. The supernatant was then removed and 200 μL of dimethyl sulfoxide (DMSO) was added. Optical density at 450 nm was measured using a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Migration and invasion assays
Migration and invasion assays were performed using a 24-well Transwell chamber (8 μm; Costar, Boston, MA, USA) with or without Matrigel (BD Biosciences, San Jose, CA, USA). After transfection, cells in serum-free media were added into upper chamber. Media containing 10% FBS were added into the bottom chamber. The setup was incubated in the 37°C humidified CO2 for 48 h. Cells that passed through the filter were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. The number of migratory and invasive cells was counted in five randomly selected high-power fields under a microscope.
Measurement of apoptosis
Apoptosis was assayed using Nucleosome ELISA kit (Merck Millipore, Billerica, MA, USA) according to previous reports.13 After transfection, the cell lysate and the biotinylated detector antibody were added (in triplicate) in the 96-well plate coated with DNA-binding protein at a cell density of 1×106 per well. Then, mono-and oligonucleosomes were captured on pre-coated DNA-binding proteins. The anti-histone 3 (H3) biotin-labeled antibody was then bound to the histone component of captured nucleosomes and was detected following incubation with streptavidin-linked horseradish peroxidase (SA-HRP) conjugate. HRP catalyzed the conversion of colorless tetramethylbenzidine to blue. The addition of stop solution changed the color to yellow, the intensity of which was proportional to the number of nucleosomes in the sample. The absorbance was measured by a microplate reader (Bio-Rad Laboratories Inc.) at a wavelength of 450 nm. The induction of apoptosis was evaluated by assessing the enrichment of nucleosome in the cytoplasm and determined exactly as described in the manufacturer’s protocol.
Caspase activity assays
Caspase-3 activity was assayed using the Caspase-3 activity assay kit (Beyotime, Beijing, China) according to the manufacturer’s instructions.
Western blot
Cell lysates were obtained using RIPA lysis buffer (Beyotime), and the protein concentrations were determined by a BCA protein assay kit (Pierce, Rockford, IL, USA). 20–50 μg of the proteins were separated by SDS-PAGE gels and transferred onto a polyvinylidene fluoride (PVDF) membrane (Merck Millipore). Antibodies against caspase-3, Bcl-2, Mcl-1, Bcl-xl, E-cadherin, N-cadherin and Vimentin were obtained from Cellular Signaling Technology (Danvers, MA, USA). GAPDH was purchased from Sigma-Aldrich Co. PVDF membrane was incubated with primary antibody overnight at 4°C. Then, HRP-conjugated goat anti-rabbit and sheep anti-mouse IgG (Cellular Signaling Technology) were used as secondary antibodies, and the immunoblots were visualized by using ECL Reagents (Pierce) followed by densitometry using the ImageJ software (NIH, Bethesda, MD, USA).
Statistical analyses
Statistical analyses were performed using SPSS 16.0 (SPSS Inc., Chicago, IL, USA) or Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Data were obtained from at least three independent experiments and presented as mean ± SD. The significance of differences was determined using Student’s t-test for two groups, and one-way ANOVA was used for more than two groups. Kaplan–Meier analysis and log-rank test were used for survival analysis. P-value <0.05 was considered to be statistically significant.
Results
PVT1 was upregulated in RCC tissues and cells
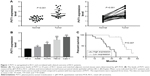
First, we measured the expression of PVT1 in 25 paired RCC (all ccRCC) tissues and adjacent normal tissues using qRT-PCR. As shown in Figure 1A, we observed significantly higher expression of PVT1 in the RCC tissues compared to normal tissues (P<0.001). Subsequently, the expression of PVT1 in RCC cells was further measured. As expected, PVT1 expression was also upregulated in RCC cells (A498, 786-O, ACHN and Caki-1) compared with human normal renal epithelial cells (HK-2) (Figure 1B). Caki-1 exhibited the highest level of PVT1, followed by 786-O cells. Thus, Caki-1 and 786-O cells were chosen for further analysis. Kaplan–Meier analysis of the 60-month follow-up survival survey demonstrated that high expression of PVT1 indicated poor survival of patients (P=0.007; Figure 1C). Next, we studied the relationship between PVT1 expression and several clinical parameters. A median value of PVT1 expression (2.53) in 25 RCC tissues was used to divide these patients into high-expression and low-expression groups. As indicated in Table 1, PVT1 was closely associated with TNM stage (P=0.007), Fuhrman grade (P=0.002), lymph node metastasis (P=0.016) and tumor size (P=0.024). However, no significant correlations were observed between PVT1 expression and other clinicopathological factors, such as gender and age. These findings provide initial evidence that aberrant expression of PVT1 might be involved in tumorigenesis and development of RCC.
  | Table 1 Correlations between clinical features and the expression of PVT1 in RCC patients |
PVT1 promotes the malignant phenotypes of RCC cells
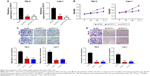
In order to elucidate the function of PVT1 in RCC progression, sh-PVT1 1 and sh-PVT1 2 were used to decrease the expression of PVT1 in 786-O and Caki-1 cells, and the transfection efficiency was tested using qRT-PCR (Figure 2A). Proliferation of 786-O and Caki-1 cells was determined in response to PVT1 knockdown by MTT assay, and results showed that the proliferation of both 786-O and Caki-1 cells was significantly repressed after silencing of PVT1 (Figure 2B). Then, transwell assay was conducted to determine the effect of downregulation of PVT1 on RCC cell invasion and migration. The data implied that silencing of PVT1 remarkably attenuated the number of migrated and invaded cells compared with control sh-NC-transfected cells (Figure 2C and D). Taken together, the abovementioned findings suggested that PVT1 might exert tumorigenic activity in RCC cells.
Downregulation of PVT1 promotes apoptosis and suppresses epithelial–mesenchymal transition (EMT) of RCC cells
Nucleosome ELISA assay was performed as described earlier to examine the effect of PVT1 on the apoptosis of RCC cells. As indicated in Figure 3A, increased apoptosis was observed in 786-O and Caki-1 cells transfected with sh-PVT1. To further confirm the effect of PVT1 on the apoptosis, the activation of caspase-3 was measured using fluorometric enzyme assay and Western blot. It was found that activity and cleavage of caspase-3 were increased after silencing of PVT1 (Figure 3B and C). It was well documented that the process of apoptosis was subjected to the regulation of Bcl-2 family protein.14,15 We detected the levels of Bcl-2 family proteins by Western blot. As shown in Figure 3C, the expression of Bcl-2, Mcl-1 and Bcl-xl was downregulated after silencing of PVT1. EMT progression is of great importance for the progression of RCC.16 Then, we attempted to explore whether PVT1 could affect the EMT progression of RCC cells. As indicated in Figure 3D, depletion of PVT1 resulted in upregulation of E-cadherin as well as downregulation of N-cadherin and Vimentin. Overall, the abovementioned findings pointed to PVT1 as a regulator of apoptosis as well as EMT.
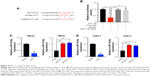
PVT1 interacts with miR-16-5p and acts as its sponge
It has been reported that there are interactions between the lncRNAs and miRNAs in the tumorigenesis of various cancers.17 To address the regulatory mechanism of PVT1, the potential targeted miRNAs of PVT1 were forecasted by bioinformatics analysis software, including Starbase and TargetScan. As a result, miR-16-5p was obtained as the candidate miRNA (Figure 4A). To verify whether PVT1 could directly interact with miR-16-5p, the full-length PVT1 or the mutant PVT without the putative binding site was cloned into the pMir-Reporter vector named PVT1-wt and PVT1-mut, respectively. The plasmids were transfected into 293 cells together with miR-16-5p mimic or the negative control. The results revealed that luciferase activity of PVT1-wt was significantly reduced by miR-16-5p mimic (Figure 4B). By contrast, the luciferase activity of PVT1-mut presented little changes (Figure 4B). In order to further verify the influence of PVT1 on miR-16-5p expression, 786-O and Caki-1 cells were transfected with pcDNA3.1-PVT1 or sh-PVT1. As shown in Figure 4C, cells with PVT1 overexpression showed significantly decreased miR-16-5p expression. Conversely, cells transfected with sh-PVT1 presented remarkably increased miR-16-5p expression (Figure 4C and D). Collectively, these findings suggested that PVT1 was a direct target of miR-16-5p, and miR-16-5p/PVT1 axis was a possible mechanism underlying the progression of RCC.
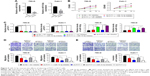
Inhibition of miR-16-5p partly reversed regulatory effects induced by downregulation of PVT1
Then, miR-16-5p inhibitor was transfected into RCC cells to repress miR-16-5p expression, which was upregulated by PVT1 downregulation. As indicated in Figure 5A, transfection of miR-16-5p inhibitor significantly repressed the expression of miR-16-5p. Furthermore, we investigated whether miR-16-5p was involved in the effects of PVT1 on proliferation, migration and invasion. As indicated in Figure 5B, transfection of miR-16-5p inhibitor partly reversed the effect of silencing of PVT1 on cell proliferation in RCC cells. Furthermore, we found that the inhibition of miR-16-5p could reduce the sh-PVT1-mediated promotive effect on miR-16-5p expression (Figure 5C). In addition, transwell analysis further confirmed that transfection of miR-16-5p also partly reversed the effect of downregulation of PVT1 on migration and invasion of RCC cells (Figure 5D and E). These findings suggested that PVT1 promoted the RCC proliferation, migration and invasion at least partly via negatively regulating the miR-16-5p.
Inhibition of miR-16-5p partly abrogates apoptosis and inhibition of EMT caused by silencing of PVT1
Furthermore, we investigated whether miR-16-5p mediated the apoptosis and inhibition of EMT induced by PVT1 downregulation on RCC cell lines. Our data indicated that overexpression of miR-16-5p inhibitor significantly attenuates the apoptosis (Figure 6A), activity of caspase-3 (Figure 6B) and cleavage of caspase-3 (Figure 6C) induced by silencing of PVT1. Moreover, the upregulation of E-cadherin and downregulation of N-cadherin and Vimentin, caused by silencing of PVT1, could be abrogated by ectopic expression of miR-16-5p inhibitor (Figure 6D). Taken together, these results further indicated that PVT1 exerts its oncogene function via negative regulation of miR-16-5p.
Discussion
LncRNAs are emerging as vital players in tumor biology. Mounting evidence indicated that lncRNAs, which may function as oncogenes or tumor suppressors, participated in the carcinogenesis and progression of various tumors including RCC.7,17 LncRNA PVT1 locates at chromosome 8q24, which is an area with frequent copy number amplification.18 A number of studies have indicated that overexpression of PVT1 was associated with tumor progression and poor survival in various cancers.19 Although it was found that high expression of PVT1 predicts unfavorable prognosis in patients with ccRCC,20 little is known about the function of PVT1 and its molecular mechanism in RCC. In the present study, we studied the pattern of PVT1 expression in RCC tissues and cell lines. Moreover, the potential prognosis significances of PVT1 in RCC patients and its effect in RCC cells were investigated.
In our study, we found that PVT1 was significantly overexpressed in RCC tissues and cell lines. More importantly, knock-down of PVT1 significantly repressed cell proliferation, migration, invasion and induced apoptosis. Our findings are similar to previous studies in which PVT1 contributed to the oncogenesis of pancreatic cancer, hepatocellular carcinoma, cervical cancer and bladder cancer.11,21–23 Collectively, these findings suggested that PVT1 may function as an oncogene in RCC and upregulation of PVT1 contributes to RCC development and progression.
Although PVT1 is involved in the progression of various cancers, little is known about the underlying mechanism. EMT is an essential process for tumor invasion and metastasis. Amounting evidence indicates that some lncRNAs can promote metastasis via regulation of critical proteins thereby triggering migration and invasion.24 In our study, hallmark proteins of mesenchymal cells such as N-cadherin and Vimentin were downregulated while hallmark protein of epithelial cell such as E-cadherin was upregulated after silencing of PVT1. Our findings are in accordance with previous studies in which PVT1 could promote EMT in pancreatic, cervical and esophageal cancer cells.25–27 Overall, these findings suggested that PVT1 promotes RCC progression via modulating the EMT process.
The mechanisms by which lncRNA exert their effects are diverse; among them the mechanism by which lncRNAs function as miRNA sponge to regulate endogenous miRNAs is important. By using bioinformatics analysis software, we identified some candidate miRNAs that might bind with PVT1, including miR-16-5p. Besides bioinformatics analysis, luciferase reporter assay also defined that miR-16-5p is the target of PVT1. Moreover, miR-16-5p inhibitor could partly reverse the regulatory effects of downregulation of PVT1. According to previous studies, miR-16-5p is downregulated and acts as a tumor inhibitor in giant cell tumor of bone.28 In addition, downregulation of miR-16-5p is associated with severe progression of gastric cancer.29 Taken together, these findings imply that miR-16-5p may function as a tumor inhibitor. To date, there is little knowledge about the role of miR-16-5p in RCC, and it would be interesting to further investigate the function of miR-16-5p in the tumorigenesis of RCC. However, noteworthy, we found that inhibition of miR-16-5p cannot fully reverse the effects of knockdown of PVT1. It indicates that PVT1 may also be acting via other molecules/pathways, and further investigation is required to test this.
Conclusion
Altogether, we have described the clinicopathological features and the function of PVT1 in RCC cell proliferation, migration, invasion, apoptosis and EMT. In this study, PVT1 acts as a molecular sponge for miR-16-5p. Our findings suggest that PVT1 may serve as a novel therapeutic target for RCC treatment. However, there are still drawbacks in our research. For example, the clinical samples are relatively small and there is lack of in vivo data. In addition, the target of miR-16-5p is not investigated in our study. In the future, we will examine the function of PVT1 in vivo and the molecular mechanisms of miR-16-5p in the development of RCC.
Acknowledgment
This study was supported by the Science Foundation of Health Bureau of Wenzhou City of Zhejiang, China (2014s0236) and Zhejiang Provincial Natural Science Foundation, China (LY16H050032).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332 | ||
Rini BI, Rathmell WK, Godley P. Renal cell carcinoma. Curr Opin Oncol. 2008;20(3):300–306. doi:10.1097/CCO.0b013e3282f9782b | ||
Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J Surg Pathol. 2003;27(5):612–624. | ||
Patil S, Ishill N, Deluca J, Motzer RJ. Stage migration and increasing proportion of favorable-prognosis metastatic renal cell carcinoma patients: implications for clinical trial design and interpretation. Cancer. 2010;116(2):347–354. doi:10.1002/cncr.24713 | ||
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006 | ||
Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127(3):761–771. doi:10.1172/JCI84424 | ||
Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634 | ||
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi:10.1016/j.tcb.2011.04.001 | ||
Xu MD, Wang Y, Weng W, et al. A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin Cancer Res. 2017;23(8):2071–2080. doi:10.1158/1078-0432.CCR-16-0742 | ||
Guo K, Yao J, Yu Q, et al. The expression pattern of long non-coding RNA PVT1 in tumor tissues and in extracellular vesicles of colorectal cancer correlates with cancer progression. Tumour Biol. 2017;39(4):1010428317699122. doi:10.1177/1010428317699122 | ||
Gao YL, Zhao ZS, Zhang MY, Han LJ, Dong YJ, Xu B. Long noncoding RNA PVT1 facilitates cervical cancer progression via negative regulating of miR-424. Oncol Res. 2017;25(8):1391–1398. doi:10.3727/096504017X14881559833562 | ||
Yu R, Yu BX, Chen JF, et al. Anti-tumor effects of Atractylenolide I on bladder cancer cells. J Exp Clin Cancer Res. 2016;35:40. doi:10.1186/s13046-016-0444-6 | ||
Kuo PL, Chiang LC, Lin CC. Resveratrol-induced apoptosis is mediated by p53-dependent pathway in Hep G2 cells. Life Sci. 2002;72(1):23–34. | ||
Cory S, Roberts AW, Colman PM, Adams JM. Targeting BCL-2-like proteins to kill cancer cells. Trends Cancer. 2016;2(8):443–460. doi:10.1016/j.trecan.2016.07.001 | ||
Garner TP, Lopez A, Reyna DE, Spitz AZ, Gavathiotis E. Progress in targeting the BCL-2 family of proteins. Curr Opin Chem Biol. 2017;39:133–142. doi:10.1016/j.cbpa.2017.06.014 | ||
Piva F, Giulietti M, Santoni M, et al. Epithelial to mesenchymal transition in renal cell carcinoma: implications for cancer therapy. Mol Diagn Ther. 2016;20(2):111–117. doi:10.1007/s40291-016-0192-5 | ||
Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108(10):1927–1933. doi:10.1111/cas.13342 | ||
Tseng YY, Moriarity BS, Gong W, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512(7512):82–86. doi:10.1038/nature13311 | ||
Cui M, You L, Ren X, Zhao W, Liao Q, Zhao Y. Long non-coding RNA PVT1 and cancer. Biochem Biophys Res Commun. 2016;471(1):10–14. doi:10.1016/j.bbrc.2015.12.101 | ||
Bao X, Duan J, Yan Y, et al. Upregulation of long noncoding RNA PVT1 predicts unfavorable prognosis in patients with clear cell renal cell carcinoma. Cancer Biomark. 2017;21(1):55–63. doi:10.3233/CBM-170251 | ||
Lan T, Yan X, Li Z, et al. Long non-coding RNA PVT1 serves as a competing endogenous RNA for miR-186-5p to promote the tumorigenesis and metastasis of hepatocellular carcinoma. Tumour Biol. 2017;39(6):1010428317705338. doi:10.1177/1010428317705338 | ||
Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. 2018;233(5):4044–4055. doi:10.1002/jcp.26072 | ||
Zhuang C, Li J, Liu Y, et al. Tetracycline-inducible shRNA targeting long non-coding RNA PVT1 inhibits cell growth and induces apoptosis in bladder cancer cells. Oncotarget. 2015;6(38):41194–41203. doi:10.18632/oncotarget.5880 | ||
Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139(2):269–280. doi:10.1002/ijc.30039 | ||
Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25(7):637–644. doi:10.1080/1061186X.2017.1307379 | ||
Wu BQ, Jiang Y, Zhu F, Sun DL, He XZ. Long noncoding RNA PVT1 promotes EMT and cell proliferation and migration through downregulating p21 in pancreatic cancer cells. Technol Cancer Res Treat. 2017:1533034617700559. | ||
Zheng X, Hu H, Li S. High expression of lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2016;12(4):2357–2362. doi:10.3892/ol.2016.5026 | ||
Sang S, Zhang Z, Qin S, Li C, Dong Y. MicroRNA-16-5p inhibits osteoclastogenesis in giant cell tumor of bone. Biomed Res Int. 2017;2017:3173547. doi:10.1155/2017/3173547 | ||
Zhang J, Song Y, Zhang C, et al. Circulating miR-16-5p and miR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics. 2015;5(7):733–745. doi:10.7150/thno.10305 |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.