Back to Journals » International Journal of General Medicine » Volume 14
LncRNA PRNCR1 rs1456315 and CCAT2 rs6983267 Polymorphisms on 8q24 Associated with Lung Cancer
Authors Yu WL, Yao JJ, Xie ZZ, Huang YJ, Xiao S
Received 12 November 2020
Accepted for publication 18 December 2020
Published 25 January 2021 Volume 2021:14 Pages 255—266
DOI https://doi.org/10.2147/IJGM.S290997
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Wei-Ling Yu,1,2,* Jin-Jian Yao,2,3,* Zong-Zhou Xie,1 Yan-Jing Huang,4 Sha Xiao5
1Oncology Department of Haikou City People’s Hospital, Haikou 570208, Hainan, People’s Republic of China; 2Key Laboratory of Emergency and Trauma, Ministry of Education of Hainan Medical University, Haikou 571199, Hainan, People’s Republic of China; 3Emergency Center of Hainan General Hospital Affiliated to Hainan Medical University, Haikou 570311, Hainan, People’s Republic of China; 4Oncology Department of Hainan General Hospital Affiliated to Hainan Medical University, Haikou 570311, Hainan, People’s Republic of China; 5School of Public Health of Hainan Medical University, Haikou 571199, Hainan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Sha Xiao
School of Public Health of Hainan Medical University, Haikou 571199, Hainan, People’s Republic of China
Tel +86-18808977719
Email [email protected]
Background: Long noncoding RNA single nucleotide polymorphisms (lncRNA-SNPs) PCAT1 rs710886, PRNCR1 rs1456315 and CCAT2 rs6983267 on 8q24 region present generalizability in the susceptibility to multiple cancers, however, the influence of rs710886, rs1456315 and rs6983267 on lung cancer has not been assessed. The aim of this study was to investigate associations between three lncRNA-SNPs and lung cancer.
Methods: A case–control study was performed on 438 patients with lung cancer and 456 healthy controls in the Han population from southern China. The collected samples were genotyped by the TaqMan genotyping, and the association with clinical characteristics, including age, gender, drinking status, smoking status, pathological types and clinical stages were analyzed. And the SNP function prediction was based on lncRNASNP2, RNAfold and GTEx.
Results: The rs1456315 T allele increased the risk of lung cancer [OR=1.95, 95% CI (1.58– 2.43), P=0.003] compared to the rs1456315 C allele, and rs1456315 significantly increased the risk of lung cancer in the dominant model [OR=1.86, 95% CI (1.16– 3.00), P=0.002]. The rs6983267 G allele, compared with the T allele, increased the risk of lung cancer [OR=1.29, 95% CI (1.07– 1.57), P=0.007], and rs6983267 was identified as a risk factor for lung cancer [OR=1.28, 95% CI (1.06– 1.55), P=0.003] in the additive model. Both rs1456315 and rs6983267 demonstrated significance after adjusting for the smoking status, drinking status and age. The structure prediction found rs6983267 and rs1456315 influence the secondary structure of its lncRNA. The results from lncRNASNP2 indicated that rs6983267 and rs1456315 change gain/loss target of miRNAs.
Conclusion: PRNCR1 rs1456315 and CCAT2 rs6983267 on 8q24 region are significantly associated with lung cancer in the Han population of southern China and alter the potential biological function in bioinformatic analysis, and the results further extended generalism of the susceptibility of cancer-associated lncRNA-SNPs to lung cancer and underlying mechanism involved in lung cancer.
Keywords: prostate cancer-associated transcript 1, prostate cancer noncoding RNA 1, colon cancer-associated transcript 2, long noncoding RNAs, single nucleotide polymorphism
Introduction
Lung cancer has the highest incidence and mortality of all malignant tumors, accounting for 11.6% of the total cases of cancer and 18.4% of the total cancer-related deaths.1 Environment factors include smoking, alcohol, age, air pollution and life style have been identified to modulate risk of cancer, and the important risk factor of cigarette smoking accounts for approximately 80% of lung cancer patients, suggesting the individual genetic factors may influence susceptibility to lung cancer. Additionally, accumulative evidence suggested that cancer-associated single nucleotide polymorphism (SNP) on chromosome 8q24 bears susceptibility in common.2–7 Genome-wide association studies have consistently identified multiple independent regions on chromosome 8q24 associated with cancers, and SNPs in lncRNA sequence on 8q24 region also share a predisposition to cancers,7,8 LncRNA-SNPs, SNPs in the LncRNA sequence, represent an attractive class of markers that illustrate association and modify lncRNA structure and function with a predictive biological significance. The cancer-associated lncRNA-SNP locus could alter lncRNA epigenetics in terms of allele-specific regulation and functionally modulate allele-specific interactions between miRNA: lncRNA and lncRNA secondary structural change.
It has been reported that lncRNA-SNPs on chromosome 8q24 demonstrated association with cancers,4–7 the prostate cancer-associated transcript 1 (PCAT1) gene lncRNA-SNP rs710886 (A>G) is correlated with the risk of bladder cancer4 and lung squamous cell carcinoma,5 the prostate cancer noncoding RNA 1 (PRNCR1) gene lncRNA-SNP rs1456315 (T>C) is associated with prostate cancer, colorectal cancer5 and breast cancer,6 and the colon cancer-associated transcript 2 (CCAT2) gene lncRNA-SNP rs6983267 (T >G) is closely linked to colon cancer,8 bladder cancer9 and hepatocellular carcinoma.10 However, the correlation between these cancer-associated lncRNA-SNPs (rs710886, rs1456315 and rs6983267) and lung cancer has not yet been assessed. A case–control study was performed to explore correlations between the lncRNA-SNPs and lung cancer, and analyze its potential function.
Materials and Methods
Study Population
All the participants were self-reported as unrelated Han ethnicity from Hainan Province. A total of 438 patients with lung cancer were enrolled. The enrolled patients were histologically diagnosed with lung cancer between March 2011 and July 2017 at Haikou City People’s Hospital and Hainan General Hospital affiliated to Hainan Medical University. A total of 456 persons who came to the hospitals for a routine physical examination during the same time period were recruited as healthy controls, and healthy controls with a history of respiratory diseases and evidence of malignancy were excluded.
Data Collection
The characteristics of each subject, including age, sex, smoking status, drinking status, and history of diabetes, were collected via a questionnaire. The classification of smokers was as follows: never-smokers were defined as those who had never smoked in their lifetime; ever-smokers were defined as those who smoked regularly before the date of lung cancer diagnosis; and smokers were defined as those who smoked currently at the date of completion of the questionnaire. In the same manner, drinkers were classified as nondrinkers, ever-drinkers, and drinkers. The clinical features of lung cancer were collected from the patients’ medical records. Clinical data were analyzed according to the eighth edition of the lung cancer stage classification.11
The study was approved by the Hainan Medical University Ethics Committee in accordance with the Declaration of Helsinki. Written informed consent was obtained from patients or their assigned relatives due to the patients are unconscious at emergency department or intensive care unit in affiliated hospitals of Hainan Medical University (IRB:HYLL-2019-034).
Peripheral Blood DNA Extraction and Genotyping
Genomic DNA was isolated from whole blood collected in EDTA tubes using an AxyPrep Blood Genomic DNAMiniprep Kit (Axygen Biosciences, CA, US) according to instructions provided by the manufacturer. DNA yield and purity were qualified by ultraviolet light spectroscopy for DNA analysis.
Genotyping was performed with TaqMan SNP Genotyping Assays (Applied Biosystems, California, USA) on the following SNPs (gene, SNP, ABI identifier assay mix): PCAT1, rs710886, C__753164210; PRNCR1, rs1456315, C__7531200_20; and CCAT2, rs6983267, C__29086771_20. The conditions used for TaqMan genotyping were as follows: 2× TaqMan genotyping master mix (ABI) 2.5 μL; 20× SNP genotyping assay mix (ABI) 0.25 μL; and template DNA 2.25 μL. The PCR conditions were 95°C for 10 min, denaturation at 95°C for 15 s, and 60°C for 90 s. A total of 40 cycles were completed.
Genotypic Function Exploration by Web-Based Bioinformatics
We used RNAfold (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi) and LncRNASNP2 (http://bioinfo.life.hust.edu.cn/lncRNASNP/) database to predict the biological effect of the significant SNPs on lncRNAs. RNAfold is a classic database to predict RNAs structure and energy change in RNA formation according to RNA sequence. Free energy represents the energy requires to change the secondary structure from the current RNA structure. LncRNASNP2 provides comprehensive resources of SNPs in lncRNAs, including SNP effects on lncRNA structure and lncRNA-miRNA binding interaction. And we further explored the effects of SNPs on gene expression by investigating a public database of GTEx portal (http://www.gtexportal.org/home/).
Statistical Analysis
Statistical analysis was performed using STATA 10.0 SE. The characteristics of the case and control groups were compared by the Χ2 test and the Wilcoxon rank-sum test if necessary. The Hardy–Weinberg equilibrium (HWE) test was performed by the likelihood ratio Χ2 test. Chi-square analysis was applied to examine differences in allelic and genotypic distribution, the Cochran–Armitage test was used for genotypic trend analysis, and genetic models fit genetic susceptibility. Multinomial logistic regression was employed to estimate the odds ratio (OR) and 95% confidence interval (CI). All P-value <0.05 was considered significant.
The false-positive report probability (FPRR) was calculated as described previously,12 only the significant result with an FPRP value less than 0.2 was considered a noteworthy finding. The threshold set 0.2 and assigned a prior probability of 0.1 to detect an odds ratio (OR) of 0.67/1.5 (protective/risk effects) for an association. We also chose P<0.05 as a standard of statistical significance.
Results
Characteristics of the Participants
As listed in Table 1, the median age in the case group was older than that in the control group (P<0.001), while there was no significant difference in sex between the two groups. Nonsmokers exhibited a decreased incidence of lung cancer, and ever-smokers, smokers, and drinkers were statistically more common in the case group than in the healthy control group.
 |
Table 1 Characteristics of Lung Cancer Cases and Controls |
Hardy–Weinberg (HWE) Analysis
rs710886 (98.9%), rs1456315 (99.6%) and rs6983267 (99.3%) showed successful genotyping rates. The HWE test showed no significant difference in SNPs in the control group (rs710886: Χ2=0.009, P=0.927; rs1456315: Χ2=3.49, P=0.062; rs6983267: Χ2=0.198, P=0.657), indicating that control samples were obtained from the same population.
Associations of rs710886, rs1456315, and rs6983267 with Lung Cancer
The allelic and genotypic distributions of the genotypes and associations between the SNPs and lung cancer are summarized in Table 2. The rs1456315 T allele significantly increased the risk of lung cancer compared to the C allele, and the rs1456315 TT genotype was associated with lung cancer in both the additive and dominant models; however, there was no significant correlation between the rs1456315 genotype and pathological type or clinical stage as illustrated in Table 3. The rs6983267 G allele significantly increased the risk of lung cancer compared with the T allele. The genotypic analysis found that rs6983267 was significant in both the additive and recessive models and associated positive lymph node; however, there was no relationship between rs6983267 and the pathological type or clinical stage as listed in Table 4, and no significant relationship was found between the risk of lung cancer and the rs710886 genotype.
 |
Table 2 Association of rs710886, rs1456315, and rs6983267 with Lung Cancer |
 |
Table 3 Clinicopathologic Characteristics of Lung Cancer Patients Stratified by Polymorphic Genotypes of rs1456315 |
 |
Table 4 Clinicopathologic Characteristics of Lung Cancer Patients Stratified by Polymorphic Genotypes of rs6983267 |
LncRNA-SNPs rs710886, rs1456315, and rs6983267 and High-Risk Factors in the Multinomial Logistic Regression Analysis
The effects of rs1456315, rs6983267, age, smoking status, drinking status, type 2 diabetes and sex on lung cancer were evaluated by multinomial logistic regression analysis. The results demonstrated that rs1456315, rs6983267, smoking status, drinking status, and age were significantly associated with lung cancer, as shown in Table 5, indicating that the rs1456315 and rs6983267 genotypes were still associated with lung cancer after adjusting for the smoking status, drinking status, and age.
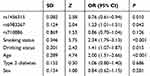 |
Table 5 SNPs and Risk Factors in Multinomial Logistic Regression Analysis |
Stratification Analysis of High-Risk Factors for Lung Cancer
In the stratification analysis, we redefined ever-smokers and smokers as smokers and nonsmokers as controls, and ever-drinkers and drinkers as drinkers and nondrinkers as controls. The analysis revealed in Table 6, the rs1456315-TT significantly increased the risk of lung cancer in smokers, nondrinkers and patients older than 50 years, and the rs6983267-GG increased the risk of lung cancer in smokers and drinkers as shown in Table 7.
 |
Table 6 Stratification Analysis of rs1456315 in the Case and Control Groups Based on the Smoking Status, Drinking Status and Age |
 |
Table 7 Stratification Analysis of rs6983267 in the Case and Control Groups Based on the Smoking Status, Drinking Status and Age |
False Positive Report Probability (FPRP) Analysis
The FPRP values for significant findings at different prior probability levels are illustrated in Table 8, as defined standard that assigned a prior probability of 0.1 to detect an odds ratio (OR) of 0.67/1.5 (protective/risk effects), the evident association for rs1456315 remained noteworthy in allelic comparison (T vs C), additive model (TT/TC/CC), dominant model (TT+TC vs CC) as well as genotypic comparison in subgroup of nondrinker between lung cancer and controls. And for rs6983267, allelic comparison (T vs G), additive model (TT/TG/GG) as well as genotypic comparison in subgroup of smokers between lung cancer and controls demonstrated significantly associations.
 |
Table 8 False-Positive Report Probability (FPRP) Values for the Significant Findings |
Function Exploration Based on Bioinformatic Database
We use LncRNASNP2 and RNAfold to predict the function of rs1456315 (T>C) and rs6983267 (T>G), LncRNASNP2 used to explored interaction between miRNA and lncRNA. The rs1456315 in LncRNA PRNCR1 (NONHSAT216393.1) may bring gain target of hsa-miR-376a-2-5p and loss target of hsa-miR-3149[Table S1 and Figure S1]. The rs6983267 in LncRNA CCAT2 (NONHSAT216396.1) might indicate gain target of hsa-miR-6820-3p, hsa-miR-627-3p, hsa-miR-5190, hsa-miR-4276, hsa-miR-3164, while loss target of hsa-miR-519e-3p, hsa-miR-371a-3p, hsa-miR-33b-3p, hsa-miR-515-3p[Table S2 and Figure S2].
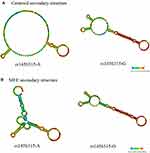
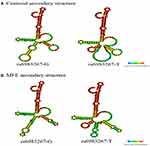
In RNAfold analysis, The centroid secondary and minimum free energy (minimum free energy, MFE) structure of rs1456315 and rs6983267 are shown in Figures 1 and 2, respectively, The genotype of rs1456315 also bring changes to centroid secondary and MFE of the thermodynamic ensemble, and the MFE of the thermodynamic ensemble was alter from −23.17kcal/mol (rs1456315-A) to −27.29kcal/mol (rs1456315-G), indicating rs1456315 may change the structural stability of lncRNA PRNCR1. And the rs6983267 in LncRNA CCAT2 would change the secondary structure of lncRNA, moreover, the MFE of the thermodynamic ensemble was change from −45.3kcal/mol (rs6983267-G) to −46.47kcal/mol (rs6983267-T), which suggests rs6983267-T may increase the structural stability of lncRNA CCAT2, comparing to rs6983267-G.
In expression analysis for rs1456315 (T>C) and rs6983267 (T>G), we found that rs6983267 contribute to expression quantitative trait loci (eQTL) based on the public database GTEx Portal. rs6983267-GG upregulated the expression CASC8 (cancer susceptibility 8, minus strand) in whole blood (Median value GG:0.409; GT:-0.071;TT:-0.3567, P= 6.3e-10) [Figure S3], while rs1456315 have no found related information, we further explored rs1456315 in its haplotype, including the tag-SNPs of rs146315, rs7007694, 7841060, rs16901946, rs1016343, rs13252298, rs13254738 and rs6983561, there are no related information available.
Discussion
This study investigated the association of cancer-associated lncRNA-SNPs (rs710886, rs1456315 and rs6983267) with lung cancer. The major finding was that the lncRNA-SNP PRNCR1 rs1456315 and lncRNA CCAT2 rs6983267 were correlated with lung cancer in the Han population of southern China, the stratification study shows rs1456315-TT significantly increased the risk of lung cancer in smokers, nondrinkers and patients older than 50 years, and rs6983267-GG increased the risk of lung cancer in smokers and drinkers, which addressed the significance of cancer-associated lncRNA-SNPs in lung cancer. In further bioinformatic exploration, we found rs6983267 and rs1456315 may influence the secondary structure of LncRNA and affect the bind to those miRNAs, rs6983267 alter the expression in eQTL on the GTEx analysis.
The rs1456315 genotype led to increased susceptibility of lung cancer. rs1456315, which is located in the 8q24 gene desert region, exerts susceptibility to various cancers, including prostate cancer, in both Japanese13 and Iranian14 populations, and is related to breast cancer6 and colorectal cancer.15 The high-risk genotype combination (TT+TC) presented a statistically significant relation to lung cancer in our study, similar to that observed in prostate cancer. Further stratification analysis of the risk factors revealed that smokers, patients older than 50 years, and nondrinkers were at increased susceptibility to lung cancer, although age and the smoking status are commonly recognized as high-risk factors for lung cancer. Interestingly, nondrinkers showed an increased risk of lung cancer. This result is consistent with the previous study that light drinking was associated with a decreased incidence of lung cancer,16 therefore, the further stratification of the drinking status is required to clarify the amount needed to decrease the risk of lung cancer.
rs1456315 exerts strong linkage disequilibrium with rs7463708 and rs72725879 (r2>0.8) based on HaploReg 4.1, indicating that the rs1456315 genotype is identical to the rs7463708 and rs72725879 genotypes, thus extending the functional significance of rs1456315. The T allele of rs1456315 can increase the gene expression of PRNCR1, which is involved in prostate carcinogenesis, possibly by changing androgen receptor affinity.4 SNPs in 8q24 have been reported to alter the secondary structure of PRNCR1 mRNA and the stability of mRNA conformation,17 therefore rs1456315 actively involved the process of PRNCR1 regulation might also occur in lung cancer.
rs6983267 is located at the conserved region of 8q24.21, and the lncRNA CCAT2 rs6983267-GG genotype was reported to have a reproducible association with colon cancer,18 cervical cancer19 and lung cancer.20 The rs6983267 of lncRNA CCAT2 is transcribed with G (CCAT2-G) or T (CCAT2-T) in a 1.7-kb RNA transcript. The allelic difference causes changes in RNA secondary structures. Changes in secondary structures can regulate the affinity of the cleavage factor I complex, and CCAT2 alters the cleavage site, thus RNA has alternative fragments with different biological functions.21
rs6983267-GG increases the expression of CCAT2, and high CCAT2 expression is related to lung adenocarcinoma susceptibility and a poor response to cisplatin chemotherapy.22 Additionally, CCAT2 rs6983267-TT genotype is associated with a decreased incidence of lung adenocarcinoma.23 Based on the theory that T/G is a protective/risk allele, rs6983267-GG also assume a risk allele for lung adenocarcinoma. And in expression quantitative trait locus analysis of whole blood, rs6983267-GG increased expression of CASC8 (also name CCAT) in whole blood, thus, taken together, rs6983267 increase likelihood to influence expressive modulation.
The false-positive report probability (FPRP) is to assess the credibility of these positive results. After FPRP analysis, the significance showed the power that further confirmed the association of rs6983267 and rs1456315 with lung cancer.
The bioinformatic analysis indicated that both rs6983267 and rs1456315 may influence lncRNA binding to miRNAs. The rs6983267 might indicate gain target of hsa-miR-6820-3p, hsa-miR-627-3p, hsa-miR-5190, hsa-miR-4276, hsa-miR-3164, while loss target of hsa-miR-519e-3p, hsa-miR-371a-3p, hsa-miR-33b-3p, hsa-miR-515-3p. Some of them involved in tumorigenesis in current literatures, the downregulation of hsa-miR-627-3p promote osteosarcoma cell proliferation and metastasis;24 miR-33b-3p also altered the cisplatin sensitivity of cancer cells by impairing the DNA damage response;25 miR-371a-3 currently reported biomarker for germ cell tumors (GCT), and clinical value of plasma miR-371a-3p level in chemotherapy naive GCT patients is a biomaker to initiate first line of chemotherapy and predict prognosis,26,27 so the rs6983267 might regulate interaction between miR-371a-3 and lncRNA CCAT2 to alter GCT diagnosis and therapy sensitivity; miR-515-3p was markedly overexpressed in individuals with gastric carcinoma compared with that in normal gastric cells (NCs) and the surgery group (P < 0.0001), and yielded an increase area under the curve (AUC) value with miR-515-3p in model construction.28 The rs1456315 may bring gain target of hsa-miR-376a-2-5p and loss target of hsa-miR-3149, and hsa-mir-3149 have reported to play important role in DNA repair and immunity by inhibiting expression of ovarian tumor protease deubiquitinase 5 (OTUD5), as a member of the ovarian tumor protease family,29 therefore rs1456315 and rs6983267 may influence binding between LncRNA and miRNA to modulate epigenetic process. Additionally, rs1456315 and rs6983267 characterize genotypic stability of LncRNA PRNCR1 and lncRNA CCAT2 secondary structure. Combined with our findings, those available evidence could shed light on the possible molecular mechanism of lung cancer.
Although we detected associations between SNP-lncRNAs and lung cancer, the identification of additional genetic risk factors would add genetic predispositions to lung cancer. There were some limitations to this study. First, prospective cohort studies are still needed to uncover the prognostic significance between SNP-lncRNAs and lung cancer incidence. Second, the study subjects were limited to the Han Chinese ethnicity, and the sample size was moderate and from two centers; therefore, large-scale studies in multiple centers across different ethnic populations are required to validate our results.
The lncRNA PRNCR1 locus rs1456315 and lncRNACCAT2 locus rs6983267 polymorphisms associated with lung cancer in the Han population of southern China, the SNP-lncRNAs may contribute to the structure and function alteration of lncRNA PRNCR1 and lncRNACCAT2, thus, the SNP-lncRNAs can be used as functional genetic markers for lung cancer and its underlying mechanism.
Acknowledgments
The authors thank the patients, their relatives and healthy subjects who willingly participated in this study. Wei-Ling Yu and Jin-Jian Yao are co-first authors for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
Foundation project: supported by the National Natural Science Foundation of China (No: 81660550); Hainan Natural Science Foundation (No: 310150); and Hainan Medical University cultivation Foundation (HYPY201911).
Disclosure
The authors state that they have no conflicts of interest to disclose.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Hu ZB, Chen JP, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118(7):2600–2608. doi:10.1172/JCI34934
3. Lin YD, Ge YQ, Wang YY, et al. The association of rs710886 in lncRNA PCAT1 with bladder cancer risk in a Chinese population. Gene. 2017;627(5):226–232. doi:10.1016/j.gene.2017.06.021
4. Bi YH, Cui ZG, Zhou BS, Yin ZH. Polymorphisms in long noncoding RNA-prostate cancer-associated transcript 1 are associated with lung cancer susceptibility in a Northeastern Chinese population. DNA Cell Biol. 2019;38(11):1357–1365. doi:10.1089/dna.2019.4834
5. Li LJ, Sun RF, Liang YD, et al. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of colorectal cancer. J Exp Clin Cancer Res. 2013;32(1):104–111. doi:10.1186/1756-9966-32-104
6. Xu T, Hu XX, Liu XX, et al. Association between SNPs in long non-coding RNAs and the risk of female breast cancer in a Chinese population. J Cancer. 2017;8(7):1162–1169. doi:10.7150/jca.18055
7. Kasagi Y, Oki E, Ando K, et al. The expression of CCAT2, a novel long noncoding RNA transcript, and rs6983267 single-nucleotide polymorphism genotypes in colorectal cancers. Oncology. 2017;92(1):48–54. doi:10.1159/000452143
8. Yeager M, Orr N, Hayes R, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi:10.1038/ng2022
9. Yadav S, Chandra A, Kumar A, Mittal B. Association of TERT-CLPTM1L and 8q24 common genetic variants with gallbladder cancer susceptibility and prognosis in North Indian population. Biochem Genet. 2018;56(4):267–282. doi:10.1007/s10528-018-9843-z
10. Wu ER, Hsieh MJ, Chiang WL, et al. Association of lncRNA CCAT2 and CASC8 gene polymorphisms with hepatocellular carcinoma. Int J Environ Res Public Health. 2019;16(16):2833–2844. doi:10.3390/ijerph16162833
11. Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017;15(1):193–203. doi:10.1016/j.chest.2016.10.010
12. Wacholder S, Chanock S, Garcia-Closas M, Ghormli LE, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–442. doi:10.1093/jnci/djh075
13. Chung S, Nakagawa H, Uemura M, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102(1):245–252. doi:10.1111/j.1349-7006.2010.01737.x
14. Sattarifard H, Hashemi M, Hassanzarei S, Narouie B, Bahari G. Association between genetic polymorphisms of long non-coding RNA PRNCR1 and prostate cancer risk in a sample of the Iranian population. Mol Clin Oncol. 2017;7(6):1152–1158. doi:10.3892/mco.2017.1462
15. AlMutairi M, Parine NR, Shaik JP, et al. Association between polymorphisms in PRNCR1 and risk of colorectal cancer in the Saudi population. PLoS One. 2019;14(9):e0220931. doi:10.1371/journal.pone.0220931
16. Choi YJ, Myung SK, Lee JH. Light alcohol drinking and risk of cancer: a meta-analysis of cohort studies. Cancer Res Treat. 2018;50(2):474–487. doi:10.4143/crt.2017.094
17. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi:10.1016/j.cell.2011.03.014
18. Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23(9):1446–1461. doi:10.1101/gr.152942.112
19. Łaźniak S, Lutkowska A, Wareńczak-Florczak. Ż, et al. The association of CCAT2 rs6983267 SNP with MYC expression and progression of uterine cervical cancer in the Polish population. Arch Gynecol Obstet. 2018;297(5):1285–1292. doi:10.1007/s00404-018-4740-6
20. Zhang X, Chen Q, He C, et al. Polymorphisms on 8q24 are associated with lung cancer risk and survival in Han Chinese. PLoS One. 2012;7(7):e41930. doi:10.1371/journal.pone.0041930
21. Redis RS, Vela LE, Lu WQ, et al. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell. 2016;61(4):520–534. doi:10.1016/j.molcel.2016.01.015
22. Gong WJ, Yin JY, Li XP, et al. Association of well-characterized lung cancer LncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumor Biol. 2016;37(6):8349–8358. doi:10.1007/s13277-015-4497-5
23. Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumor Biol. 2014;35(6):5375–5380. doi:10.1007/s13277-014-1700-z
24. He M, Shen P, Qiu C, Wang J. miR-627-3p inhibits osteosarcoma cell proliferation and metastasis by targeting PTN. Aging. 2019;11(15):5744–5756. doi:10.18632/aging.102157
25. Xu S, Huang H, Chen Y-N, et al. DNA damage responsive miR-33b-3p promoted lung cancer cells survival and cisplatin resistance by targeting p21 WAF1/CIP1. Cell Cycle. 2016;15(21):2920–2930. doi:10.1080/15384101.2016.1224043
26. Spiller CM, Lobo J, Boellaard WPA, Gillis AJM, Bowles J, Looijenga LHJ. CRIPTO and miR-371a-3p are serum biomarkers of testicular germ cell tumors and are detected in seminal plasma from azoospermic males. Cancers (Basel). 2020;12(3):760. doi:10.3390/cancers12030760
27. Mego M, van Agthoven T, Gronesova P, Chovanec M. Clinical utility of plasma miR-371a-3p in germ cell tumors. J Cell Mol Med. 2019;23(2):1128–1136. doi:10.1111/jcmm.14013
28. Han X, Xing L, Zhao H, et al. Serum miR-515-3p, a potential new RNA biomarker, is involved in gastric carcinoma. J Cell Biochem. 2019;120(9):15834–15843. doi:10.1002/jcb.28854
29. Bai M, Che Y, Lu K, Fu L. Analysis of deubiquitinase OTUD5 as a biomarker and therapeutic target for cervical cancer by bioinformatic analysis. PeerJ. 2020;8:e9146. doi:10.7717/peerj.9146
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.