Back to Journals » Cancer Management and Research » Volume 12
lncRNA PCBP1-AS1 Aggravates the Progression of Hepatocellular Carcinoma via Regulating PCBP1/PRL-3/AKT Pathway
Authors Luo T , Gao Y , Zhangyuan G, Xu X, Xue C, Jin L, Zhang W, Zhu C, Sun B , Qin X
Received 13 February 2020
Accepted for publication 10 June 2020
Published 6 July 2020 Volume 2020:12 Pages 5395—5408
DOI https://doi.org/10.2147/CMAR.S249657
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Tianping Luo,1 Yuan Gao,1 Guangyan Zhangyuan,2 Xiaoliang Xu,3 Cailin Xue,1 Lei Jin,1 Wenjie Zhang,3 Chunfu Zhu,1 Beicheng Sun,3 Xihu Qin1
1Department of Hepatobiliary Surgery, The Affiliated Changzhou NO. 2 People’s Hospital of Nanjing Medical University, Changzhou, Jiangsu Province 213164, People’s Republic of China; 2Liver Transplantation Center of the First Affiliated Hospital, Nanjing Medical University, Nanjing, Jiangsu Province 210029, People’s Republic of China; 3Department of Hepatobiliary Surgery, The Affiliated Drum Tower Hospital of Nanjing University Medical School, Nanjing, Jiangsu Province 210008, People’s Republic of China
Correspondence: Xihu Qin
Department of Hepatobiliary Surgery, The Affiliated Changzhou NO. 2 People’s Hospital of Nanjing Medical University, Changzhou, Jiangsu Province 213164, People’s Republic of China
Email [email protected]
Background: Hepatocellular carcinoma (HCC) is a very belligerent primary liver tumor with high metastatic potential. Aberrant expression of lncRNAs drives tumorous invasion and metastasis. Whether lncRNAs engage mechanisms of liver cancer metastasis remains largely unexplored.
Patients and Methods: We collected HCC tissues from the tumors and their adjacent normal samples in the Chinese population and analyzed the levels of lncRNAs by microarray analysis. The gain- and loss-of-function analysis demonstrated that PCBP1-AS1 accelerated tumorous growth and metastasis in vivo and in vitro. Moreover, we used RNA-pulldown assay to show that PCBP1-AS1 physically interacted with polyC-RNA-binding protein 1 (PCBP1); meanwhile, PCBP1-AS1 was indeed detected in RIP with the PCBP1 antibody. Mechanistically, we first explored the relationship between PCBP1‐AS1 and PCBP1 in HCC cell lines.
Results: Here we show that PCBP1-AS1, identified by microarray analysis on pre- and post-operative HCC plasma specimens, was highly expressed in human HCC, clinically verified as a prometastatic factor and markedly associated with poor prognosis in patients with hepatocellular carcinoma. PCBP1‐AS1 was negatively related with PCBP1 at the messenger RNA and protein expression levels. PCBP1-AS1 triggered PRL-3 and AKT in HCC tumor cells. Additionally, the double knockout of PCBP1 and PCBP1-AS1 abolished the PCBP1-AS1-induced PRL-3-AKT signalling pathway activation.
Conclusion: The upregulation of PCBP1-AS1 enhances proliferation and metastasis in HCC, thus regulating the PCBP1-PRL-3-AKT signalling pathway.
Keywords: hepatocellular carcinoma, long non-coding RNA, PCBP1-AS1, poly(C)-binding protein 1, AKT, hepatocarcinogenesis, metastasis
Introduction
Hepatocellular carcinoma (HCC), one of the most frequent and deadly cancers worldwide,1 occurs half in China.2,3 While there are several treatment strategies for HCC, including liver transplantation, liver resection, chemotherapy, and radiotherapy, no satisfactory and long-term prognosis of liver cancer is currently available.4 One challenge of HCC treatment is intrahepatic metastasis resulting in a high rate of postoperative recurrence.5,6 As a result, we find it essential to explore the mechanisms of HCC initiation and early metastasis for improved early diagnosis and prognosis.
Long noncoding RNAs (lncRNAs) represent transcripts longer than 200 nucleotides (nt) that are 50 capped and 30 polyadenylated, yet this class of transcripts shows limited coding potential.7,8 At the functional level, it is known that different mechanisms have been found in the implication of IncRNAs in biological processes. Reportedly, the mechanisms of lncRNAs include modulated gene transcription and post-transcriptional processes, controlled gene expression through chromatin structure remodelling, and adjusted protein function or localization.7,9 Particularly, IncRNAs play an irreplaceable role in every stage of metastasis from cancer cells which invade and migrate to distant organs.10 The “Flank10kb” theory shows that over 65% of IncRNAs lie within 10 kb of known protein-coding genes, and that cis-acting or trans-regulatory relationships may exist between lncRNAs and known genes.11,12 Thus, to investigate the association of location-associated lncRNAs with protein-coding genes, clinical specimens from HCC patients were assayed to screen differentially expressed lncRNAs. Finally, PCBP1-AS1, an IncRNA, has been selected as the candidate lncRNA located in the upstream of poly(C)-binding protein 1-antisense RNA 1(PCBP1) with a distance of 462bp, and presented a reverse transcription direction with PCBP1. The crucial roles of PCBP1 have been revealed in transcriptional and translational events,13 with its overexpression inhibiting PRL-3 expression and inactivating AKT.14 PRL-3 overexpression is found to be in a negative correlation with the prognosis of liver cancers,15 but the molecular mechanism of PCBP1-AS1 in cancer development and progression remains to be defined.
In the present study, we examined the expression of PCBP1-AS1, its relationship to clinicopathological characteristics and prognosis of patients, its regulative mechanisms of PCBP1, and its biological effect on HCC growth in vitro and in vivo. This study provides mechanistic insights into how antisense noncoding RNA regulates sense genes and reveals the important roles of PCBP1-AS1 in HCC.
Patients and Methods
Patient Samples and Cell Lines
Totally, 109 paired fresh HCC tissues (tumorous and adjacent normal samples) and relevant clinical information were obtained from patients undergoing hepatectomy at the First Affiliated Hospital of Nanjing Medical University (Nanjing, China) from June 2013 to July 2014. The study protocol was approved by the Institutional Ethics Committee of the First Affiliated Hospital of Nanjing Medical University, Nanjing, China. All patients had given written informed consent to participate in our study before surgery and that was conducted in accordance with the Declaration of Helsinki. All fresh tissues were collected and immediately frozen in liquid nitrogen. The diagnosis of all patients was confirmed by pathology. HepG2, Hep3B, MHCC97H, Huh7, SNU423, and SMMC-7721 human hepatoma and normal L02 cell lines were obtained from Nanjing KeyGen Biotech Co. Ltd (Nanjing, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) and 80 U/mL of penicillin sodium at 37°C in humidified air containing 5% carbon dioxide.
RNA Isolation and RT-qPCR
We used TRIzol reagent to isolate total RNAs from fresh tissue samples and cells according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). cDNAs were synthesized from the total RNA by random primers. The qRTPCR was conducted to assess the expression level of related lncRNAs (ABI 7900; Life Technologies). SurePrep Nuclear or Cytoplasmic RNA Purification Kit of Thermo Fisher Scientific (Rochester, Waltham, MA, USA) was used to extract RNAs from cytoplasm and nucleus. b-actin (ACTB) and GAPDH amplification were taken as the internal control. Table S2 shows the PCR primers applied in our study.
Ectopic Expression and Gene Silencing
For overexpression of PCBP1-AS1 and PCBP1, the full-length PCBP1-AS1 and PCBP1 were subcloned into the lentivirus vector GM-8980 (Genomeditech, Shanghai, China). Three shRNA sequences specifically targeting PCBP1-AS1, PCBP1 were cloned into the lentiviral vector GV248 (Gene, Shanghai, China) in addition to negative control shRNA with no sequence homology to human genes (provided by the same manufacturer). All shRNA sequences are shown in Table S2. We selected shRNAs specific for PCBP1-AS1, with PCBP1 having the highest efficiency for further experiments. Lipofectamine 3000 reagent was purchased from Invitrogen (Carlsbad, California, USA). HCC cells at the exponential growth phase were seeded into 6-well plates for 24 h at a density of 0.5×105 cells/mL and transfected according to the manufacturer’s instructions. All the vectors were labelled with enhanced green fluorescence protein (EGFP).
Cell Proliferation, Migration and Invasion Assay
Following the manufacturer’s instructions, we evaluated the proliferation capacity of HCC cells using cloning assay, cell-counting kit-8 (Dojindo Laboratories, Kumamoto, Japan), EDU (5-ethynyl-2′-deoxyuridine), and immunofluorescence staining (Millipore, Billerica, MA, USA). We also employed the transwell chamber (8μm pore size; Millipore) to assay cell migration and invasion. In the invasion experiment, a thin layer of ECM was dried on the membrane. The membrane pores were occluded and the non-invasive were blocked from migrating by the ECM layer. 5×103 cells were added to the upper lumen in serum‐free medium. DMEM with 10% FBS was added into the lower lumen. After 48 hrs, the cells were fixed with methanol, stained with crystal violet, and counted for migration or invasion through the membrane. A similar protocol to that in the above invasion assay was done in the migration assay, except that the ECM layer was not added to the chamber, and 2×103 cells were added per chamber.
In vivo Experiments
Female BALB/c nude mice (5-week-old) were purchased from the Animal Center of Nanjing University (Nanjing, China). In the model of subcutaneous transplantation, Lv-PCPB1-AS1 (PCBP1) cells (1×107) and Lv-Ctrl cells (1×107) were independently injected subcutaneously into the right groin and the left groin, respectively. The tumor volume was calculated every 5 days, and the mice were killed 30 days after transplantation. For tail vein xenograft model, cells (1×107 suspended in 200ul PBS) were injected into 6 mice in each group through the tail vein to determine the lung homing potential of cancer cells in vivo. We then sacrificed the mice. From one group we obtained sh-PCBP1-AS1 (PCBP1) cells, and the other sh-NC cells, both labelled with luciferase (EGFP). Using a 470-nm light source (Lightools Research, Encinitas, CA, USA), we used fluorescence to visualize the lung tumors. Five weeks later, all the mice were sacrificed, individual organs were removed and metastatic tissue was analyzed using haematoxylin and eosin (H&E) and immunohistochemical stains. Animal protocols were approved by the Animal Care and Use Committee of Nanjing Drum Tower Hospital of Nanjing University, and all animal experiments followed guidelines for ethical review of laboratory animal welfare of Nanjing medical university (http://iacuc.njmu.edu.cn).
RNA Fluorescence in situ Hybridization
We used Digoxygenin (DIG) labelled PCBP1-AS1 probes (Ribobio, Guangzhou, China) for RNA FISH. RNA-FISH was performed as previously described.16 Images were obtained using an FV1000 confocal laser microscope (Olympus).
RNA Pull-Down Assay and RIP Assay
Biotin-labelled PCBP1-AS1 transcript and its antisense were synthesized using Biotin RNA Labeling Mix and DIG RNA Labeling Kit (SP6. T7) (Roche) and purified with RNeasy Mini Kit (QIAGEN). The protein with biotin-labelled PCBP1-AS1 was pulled down with streptavidin magnetic beads (Pierce™, Invitrogen) after incubation overnight. Pull-down proteins were run on SDS-PAGE gels, and then silver staining was performed on the gel, and the specific bands were identified using mass spectrometry. RNA-binding protein immunoprecipitation (RIP) was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore) in accordance with the manufacturer’s protocol.
Western Blot
Total protein was extracted from tissues or cultured cells using RIPA containing phenylmethanesulfonylfluoride (PMSF) (Beyotime, Jiangsu, China). Equal amounts of proteins were boiled, separated on 10% SDS-polyacrylamide gel electrophoresis, transferred onto a PVDF membrane and visualized via an ECL kit (Millipore, MA, USA). Primary antibodies detecting PCBP1 (#sc-137249; Santa Cruz; 1:500), Histone H3 (#17168-1-AP; Proteintech; 1:3000), HSP90 (#13492; Abcam; 1:2000), PRL-3 (#sc-130355; Santa Cruz; 1:500), AKT1 (S473) (#ab81283; Abcam; 1:2000), p-AKT (T308) (#ab38449; Abcam; 1:2000), Akt (#9272; Cell Signaling; 1:2000), β-Actin (#3280; Abcam; 1:2000), or GAPDH (#ab8245; Abcam; 1:5000) were incubated overnight at the indicated dilutions. Secondary anti-rabbit and anti-mouse horseradish peroxidase-linked antibodies were purchased from Santa Cruz Biotechnology (CA, USA). Blots were developed using electrochemiluminescence (ECL) reagent (Millipore, Billerica, MA, USA). HSP90 or GAPDH antibody was used to normalize the quantity. Equal amounts of protein loading in each lane were confirmed using β-actin antibody. ImageJ software (NIH Image, Bethesda, MD, USA) was used to quantify the integrated density of the band.
Statistical Analysis
All experiments were performed independently in triplicate. Data analyses were carried out using SPSS version 20.0 software (SPSS, Palo Alto, CA, USA) and GraphPad Prism 5. Data are expressed as mean± S.E.M. and differences between two independent groups were assessed by two-tailed Student’s t-test and Mann–Whitney U-test. Log-rank analysis was applied in survival comparisons. Probability values of less than 0.05 were considered to be statistically significant.
Results
High Throughput Microarray Screens lncRNAs on Pre- and Post-Operative HCC
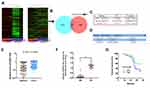
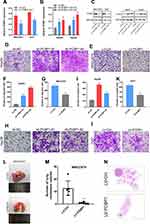
To identify transcripts that potentially change between the pre-operation and post-operation of HCC plasma specimens, we determined IncRNA expression profiles by microarray analysis. For detailed microarray data in previous studies,17 see the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under access number E-MTAB-2563. We obtained 43 lncRNA transcripts by merging lncRNA transcripts that were up-regulated preoperatively and decreased postoperatively in HCC patients (Figure 1A and B). Next, all 43 transcripts with high signal intensity (≥5), P value (≤0.01) and at least 2.5-fold were filtered to obtain 3 lncRNA candidates (Figure 1C). Among these lncRNAs, we focused on an uncharacterized lncRNA, termed PCBP1-AS1, which is located on human chromosome 2 (2p13.3). RP11-160H22.5 has been reported by Junwei Tang.17 XLOC_009752 is under way in our lab. PCBP1-AS1 was composed of eleven exons and the length of PCBP1-AS1 was 2396 base pairs (NCBI Reference Sequence: NR_033872.1) (Supplementary Figure 1A). We consulted the CPC (coding potential calculator) computational algorithm18 (Figure 1D) and CPAT (Coding Potential Assessment Tool),19 (Supplementary Figure 1B), ascertain whether the transcript of PCBP1-AS1 might encode proteins, proving that PCBP1-AS1 can encode either a protein or a small peptide. Despite its unmistakable protein-coding signatures, sequence comparisons did not determine homology with any other proteins and the classical domains/motifs using ORF Finding (http://www.ncbi.nlm.nih.gov/orffinder/). We further applied PhyloCSF to identify PCBP1-AS1 as a non-coding RNA (Supplementary Figure 1C).20 Collectively, these data demonstrated that PCBP1-AS1 had more tendency to be a non-coding RNA.
PCBP1-AS1 Is Highly Expressed in HCC and Inversely Correlated with the Prognosis of HCC Patients
PCBP1-AS1 was frequently upregulated in other tumors, which is consistent with the results of the TCGA database analysis (Supplementary Figure 1D). Moreover, the qRT-PCR analysis confirmed that PCBP1-AS1 was significantly upregulated in HCC compared with the corresponding adjacent non-cancerous liver tissues in a panel of 109 specimens (Figure 1E). Next, we determined the potential correlations between the expression of PCBP1-AS1 and clinicopathological features with clinical information in 109 tumorous tissue samples (Table 1). We classified the expression level of PCBP1-AS1 by the median value, and then divided patients into high-PCBP1-AS1 and low-PCBP1-AS1 groups. Correlation and regression analyses showed that a higher expression of PCBP1-AS1 was closely related to tumor size, vascular invasion, and metastasis (Table 1). Importantly, PCBP1-AS1 was also greatly higher in HCC patients with metastases than those without metastases (Figure 1F). Overall survival using Kaplan–Meier survival analysis showed that HCC patients with a high PCBP1-AS1 level had a significantly worse prognosis compared to those with low PCBP1-AS1 expression (Figure 1G). We performed Multivariate Cox analysis to assess its predictive value to determine the clinical outcomes of HCC patients. However, our results failed to support the high expression of PCBP1-AS1 as an independent factor for poor prognosis in HCC patients (Table S1).
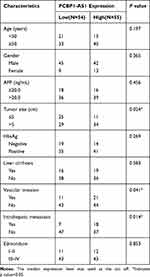 |
Table 1 Correlation Between PCBP1-AS1 Expression and Clinicopathological Characteristics of HCC Patients (n = 109) |
PCBP1-AS1 Enhances Proliferation and Metastasis of HCC Cells in vitro
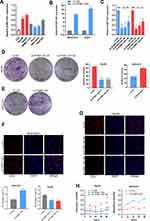
The biological function of PCBP1-AS1 was further investigated in vitro. We analyzed the expression of PCBP1-AS1 in 6 HCC cell lines and a normal human liver cell line LO2. Figure 2A shows that the expression of PCBP1-AS1 found in all HCC cell lines was comparatively increased as compared to LO2. We performed the gain- and loss-function in vitro analysis (Supplementary Figure 1E) to assess the effects of PCBP1-AS1 on cell biological behaviors. As Figure 2B shows, we screened MHCC97H and Huh7 cell lines infected with the lentivirus containing the overexpression vector of the PCBP1-AS1. Contrastingly, we knocked down PCBP1-AS1 in Hep3B and HepG2 by expressing short hairpin RNAs (shRNAs) from lentiviral vectors (Figure 2C). Colony-forming assays (Figure 2D and E), EdU (Figure 2F and G), Cell-counting kit-8 (Figure 2H), and wound healing assay (Figure 3A and B) indicated that PCBP1-AS1 significantly promoted cell viability and colony-forming ability (Figure 3C). However, the cell cycle experiments revealed that no correlation was evident between PCBP1-AS1 and cell cycle (Supplementary Figure 1F).
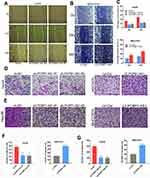
Correlation analysis between PCBP1-AS1 and clinicopathological characteristics showed that PCBP1-AS1 was highly correlated with HCC metastasis. We also conducted transwell experiments to explore whether overexpression PCBP1-AS1 has a strong effect on cell invasion and migration. Transwell assay (Figure 3D and F) (Supplementary Figure 2A-D) and matrigel assay (Figure 3E and G) (Supplementary Figure 2E-H) showed that the migration and invasion abilities were significantly increased when the PCBP1-AS1 was overexpressed in MHCC97H and Huh7 cells, while PCBP1-AS1 knockdown remarkably decreased cell migration and invasion in Hep3B and HepG2 cells (Figure 3D-G). Overall, our results clearly showed that PCBP1-AS1 has a promoting effect on migration and invasion in HCC cells.
PCBP1-AS1 Enhances HCC Growth and Metastasis in vivo
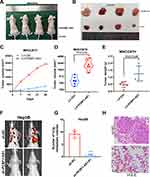
To further explore the function of PCBP1-AS1 in vivo, we established a xenotransplantation model and subcutaneously injected PCBP1-AS1 ectopic overexpressed MHCC97H cells into nude mice (Figure 4A and B). PCBP1-AS1-overexpressing xenograft tumors grew significantly faster than the control group (Figure 4C). As shown in Figure 4D and E, tumor volume and weight of PCBP1-AS1 overexpression were larger than those of LV-Ctrl cells, suggesting that PCBP1-AS1 overexpression enhanced the tumor-initiating capacity. Then, we established a model of caudal vein xenograft, in which nude mice were subcutaneously injected with tumor cells transfected with luciferase via the tail vein to observe the lung metastasis changes of HCC cells, and we measured the formation of pulmonary metastases (Figure 4F). The lung metastases ability of Hep3B cells significantly decreased under conditions of knockdown of PCBP1-AS1 in mice (Figure 4G and H). Collectively, these results demonstrated that PCBP1-AS1 overexpression promoted HCC growth and lung colonization in vivo. We next made a mechanistic study of PCBP1-AS1 in HCC.
PCBP1-AS1 Is a Direct Target of PCBP1 with Contrary Biological Behaviors
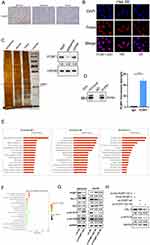
Classical antisense transcripts may preferentially control the expression of the corresponding sense transcripts.21–23 To understand the relationship between PCBP1-AS and PCBP1 in HCC, we investigated the levels of PCBP1 mRNA and protein expression in our experimental setup. The results showed that the mRNA levels of PCBP1 were significantly decreased when the PCBP1-AS1 was overexpressed in MHCC97H and Huh7 cells (Figure 5A), while its expressions were increased in Hep3B and HepG2 cells (Figure 5B). Furthermore, the expression of PCBP1 protein was detectable in PCBP1-AS1-silenced or PCBP1-AS1-overexpression cells (Figure 5C). These results suggested that PCBP1-AS1 could have an active impact on the transcriptional and post-transcriptional levels of PCBP1. To study whether the biological behaviors of PCBP1-AS1 and PCBP1 were contrary, we evaluated the migration and invasion of PCBP1 on tumor growth in vitro and vivo. PCBP1 was validated in PCBP1-silenced or sh-NC-treated and overexpressed HCC cells (Supplementary Figure 3C). Transwell assays (Figure 5D and E) showed that knockdown of PCBP1 in HepG2 cell lines sharply increased its migration and invasion abilities (Figure 5F), and a significant decrease was observed in the migration and invasion of the LV‐PCBP1‐MHCC97H cells compared to the LV‐Ctrl‐MHCC97H cells (Figure 5G). Similar results in Matrigel assays (Figure 5H and I) were achieved with Huh7 and Hep3B cells (Figure 5J and K). For a better understanding of the effects of PCBP1 on the lung homing capacity of HCC cells in vivo, we numbered the nodules in the mice injected with MHCC97H cells by way of the tail vein (Figure 5L and M). Then, the pulmonary nodules were sectioned into paraffin sections, indicating that the number of lung metastases in mice with PCBP1 overexpression was significantly lower than that of the control group, suggesting that PCBP1 overexpression significantly reduced the lung homing potential of MHCC97H cells (Figure 5N). Together, these results showed that PCBP1 overexpression decreased tumor migration, invasion, and lung colonization, which are the opposite effects of those of PCBP1-AS1 on HCC.
PCBP1-AS1 Physically Interacts with PCBP1 Protein
We sought to identify putative protein binding that interacts with PCBP1-AS1 to mechanistically describe the observed biological phenotypes. Firstly, we explored the localization of PCBP1-AS1 and PCBP1. The results of RT-PCR using isolated cytoplasmic RNA and nuclear RNA showed that PCBP1-AS1 mainly existed in the cytoplasm of Hep3B and MHCC97H cell lines (Supplementary Figure 3A), which is consistent with FISH results in Hep3B and MHCC97H cells (Figure 6B). Interestingly, ISH and IHC-P assays revealed that the strong positive signal of PCBP1 was mainly concentrated in the cytoplasm of HCC cells (Figure 6A) (Supplementary Figure 3B). Next, a biotinylated-PCBP1-AS1 protein pull-down assay was carried out in Hep3B cells. Then, we decomposed RNA associated proteins on SDS-PAGE, removed the strips and performed mass spectrometry. With a set of RNA-binding proteins screened, PCBP1 was found to strongly bind to PCBP1-AS1. PCBP1 was markedly enriched after the sense sequence of PCBP1-AS1 (Figure 6C) was pulled down, but not the antisense PCBP1-AS1 as blotted by PCBP1 antibody. RNA immunoprecipitation (RIP) was performed with extracts from the Hep3B tumor cell lines using PCBP1 as bait. We found the PCBP1-AS1 enrichment with the PCBP1 antibody against a nonspecific antibody (IgG control) (Figure 6D).
PCBP1 is Crucial to the Overexpression of PCBP1-AS1 Inducing Activation of PRL-3-AKT Signalling Pathway
The aforementioned data demonstrated that overexpression or knockdown of PCBP1-AS1 in HCC cancer cell lines could regulate PCBP1 protein levels. To better understand the functions and pathogenesis of PCBP1-AS1 in HCC, RNA sequencing was performed with HuH7-LV-NC and HuH7-LV-PCBP1-AS1. mRNA levels of PCBP1 were significantly decreased when the PCBP1-AS1 was overexpressed in Huh7 cells (Supplementary Figure 3D). Go analysis included MF, BP, and CC. Figure 6E shows that two major functions were decided for MF: heparin binding and phospholipid binding. Their functions concerning biological processes were determined to be extracellular matrix organization and homeostasis. When classified according to their respective CC classifications, the extracellular space and extracellular region made up the largest part. PCBP1-AS1 participated in such biological and cellular functions as protein digestion and absorption, ECM–receptor interaction, complement, and coagulation cascades and PI3K-Akt signaling pathway (Figure 6F). Interestingly, PCBP1-PRL-3-AKT axis may be a pathway that inhibits cancer progression, which has been demonstrated by Wang.14
PRL-3 overexpression is inversely correlated with the prognosis of HCC.15 It is one of the important mechanisms promoting cell migration, invasion, and metastasis.24,25 We then examined whether PCBP1-AS1 could influence the functions of PCBP1 that inhibit PRL-3-AKT signalling. AKT levels remained unchanged upon the silencing of PCBP1-AS1 or overexpression of PCBP1-AS1 (Figure 6G). But we detected the expression of both PRL-3 and AKT (pT308 and pSer473) in downregulated and overexpressed cell lines. Notably, a positive correlation of PCBP1-AS1 and PRL-3 with P-AKT protein expression was noted (Figure 6G). These results suggested that PCBP1-AS1 may promote the translation of PRL-3 and AKT (pT308 and pSer473) in HCC cells. Consistently, the mechanism by which PCBP1 delays transcription of PRL-3 mRNA into polyribosomes is that the 50 untranslated region (UTR) of PRL-3 mRNA has triple GCCCAG motifs, which can inhibit mRNA translation by interacting with PCBP1.14 We further examined whether the role of PCBP1 alone significantly affected PCBP1-AS1 activating PRL-3-AKT signalling. However, double knockout of PCBP1 and PCBP1-AS1 abolished the action of PCBP1-AS1 activating PRL-3-AKT signalling (Figure 6H). Thus, both PCBP1-AS1 and PCBP1 are necessary for PCBP1-PRL-3-AKT signalling.
Discussion
Death of patients with hepatocellular carcinoma results mostly from metastatic disease,26 but the mechanisms of HCC metastasis remain elusive. Previous studies show that the vital roles of aberrantly expressed lncRNAs in HCC metastasis explain the causes of most cancer-related deaths in HCC patients.27–29 Our study, using the lncRNAs microarray scanning, aimed to identify new tumor metastasis-regulated long noncoding RNAs in HCC cells and investigate the clinical significance and molecular biological function of PCBP1-AS1. We found that PCBP1-AS1 increased in hepatocellular carcinoma tissues correlated with HCC metastasis and reduced survival. The expression of PCBP1-AS1 was not merely associated with bad survival outcomes but greatly correlated with the metastasis of HCC patients as well. In vivo and in vitro experiments confirmed that PCBP1-AS1 can promote the growth and metastasis of HCC, which suggests that PCBP1-AS1 could be a potential promoter of HCC metastasis. A few coding and non-coding overlapping antisense transcripts (OATs) in eukaryotes can regulate corresponding sense transcript,30,31 and non-coding RNAs antisense can spread regulatory signals from one locus to neighbouring genes.32,33 This pattern of action has been described as other antisense transcripts, such as non-coding antisense RNA in the LncTCF7, which are engaged in the regulation of the promoter of TCF7.34 In the present study, we demonstrated that PCBP1-AS1 affects the expression of PCBP1 at the transcriptional level. Moreover, PCBP1-AS1 silencing does influence PCBP1 expression itself, and then the expression of PCBP1 downstream genes,14 PRL-3 and p-AKT. Overexpression of PCBP1-AS1 significantly increases PRL-3 and the phosphorylated active form of AKT (pT308 and pSer473) in HCC cells, while knockdown of PCBP1-AS1 sharply decreases the expression level of PRL-3 and the phosphorylated active form of AKT (pT308 and pSer473). Furthermore, analysis of lncRNA binding to protein by RNA-pulldown uncovered that PCBP1 is found to strongly bind to PCBP1-AS1.35 Previous studies have also shown that antisense RNA could regulate the expression of proteins associated with S/AS pair.36,37 In our study, PCBP1-AS1 was mainly distributed in the cytoplasm of hepatocellular carcinoma cells. The liberation of PCBP1 attenuated HCC invasion and metastasis, in clinical data as well as in vivo experiments. PCBP1-AS1 might act as a key regulator of PCBP1.38,39
PRL-3, closely associated with HCC progression, invasion, and metastasis when it is highly expressed, has a negative impact on the prognosis of HCC patients.40,41 PCBP1 can inhibit PRL-3 translation of diverse human cancers, which indicates that PCBP1 could likely be a tumor suppressor.14 We found that PCBP1-AS1 silencing did influence PCBP1 expression. Interestingly, overexpressed exogenous PCBP1-AS1 significantly increased the levels of endogenous PRL-3 protein, which also increased AKT activation; on the contrary, knock-down of endogenous PCBP1-AS1 downregulated PRL-3 protein expression and inhibited AKT activation. However, double knockout of PCBP1 and PCBP1-AS1 abolished the action of PCBP1-AS1 activating PRL-3-AKT signalling. It is thus demonstrated that PCBP1-AS1 exerts a critical role in regulating PCBP1-PRL-3-AKT signalling and can explain how PCBP1-AS1 expression regulates the expression of PCBP1. In the present experiment, PCBP1-AS1 was mainly involved in the AKT signalling pathway, which is consistent with LV-NC and LV-PCBP1-AS1 related results of RNA sequencing.
In conclusion, differences in PCBP1-AS1 expression were observed in HCC cells, and PCBP1-AS1 plays a stimulatory role in HCC tumour progression and metastasis of HCC cells by binding to PCBP1, and therefore controls HCC cell function in vitro and in vivo through regulating PCBP1-PRL-3-AKT signalling. Together, our findings suggest that PCBP1-AS1 is promising as a therapeutic target for the treatment of HCC and as a prognostic biomarker of HCC metastasis.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation (Grant Number: 81672469 to XQ), and the Social Development Foundation of Science and Technology of Jiangsu (Grant Number: BE2016658 to XQ).
Disclosure
The authors declare that there is no conflict of interest.
References
1. Margini C, Dufour JF. The story of HCC in NAFLD: from epidemiology, across pathogenesis, to prevention and treatment. Liver Int. 2016;36(3):317–324. doi:10.1111/liv.13031
2. Cao WT, Li J, Hu C, et al. Symptom clusters and symptom interference of HCC patients undergoing TACE: a cross-sectional study in China. Support Care Cancer. 2013;21(2):475–483. doi:10.1007/s00520-012-1541-5
3. Luk JM, Liu AM. Proteomics of hepatocellular carcinoma in Chinese patients. OMICS. 2011;15(5):261–266. doi:10.1089/omi.2010.0099
4. Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut. 2015;64(5):842–848. doi:10.1136/gutjnl-2014-307990
5. Lo YC, Hsu FC, Hung SK, et al. Prognosticators of hepatocellular carcinoma with intrahepatic vascular invasion. Ci Ji Yi Xue Za Zhi. 2019;31(1):40–46. doi:10.4103/tcmj.tcmj_14_18
6. Lemoine M, Thursz MR. Battlefield against hepatitis B infection and HCC in Africa. J Hepatol. 2017;66(3):645–654. doi:10.1016/j.jhep.2016.10.013
7. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi:10.1016/j.molcel.2011.08.018
8. Li Y, Egranov SD, Yang L, Lin C. Molecular mechanisms of long noncoding RNAs-mediated cancer metastasis. Genes Chromosomes Cancer. 2019;58(4):200–207. doi:10.1002/gcc.22691
9. Sanchez Y, Segura V, Marin-Bejar O, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812. doi:10.1038/ncomms6812
10. Huang Q, Yan J, Agami R. Long non-coding RNAs in metastasis. Cancer Metastasis Rev. 2018;37(1):75–81. doi:10.1007/s10555-017-9713-x
11. Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi:10.1038/nature20149
12. Joung J, Engreitz JM, Konermann S, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548(7667):343–346. doi:10.1038/nature23451
13. Huo LR, Zhong N. Identification of transcripts and translatants targeted by overexpressed PCBP1. Biochim Biophys Acta. 2008;1784(11):1524–1533. doi:10.1016/j.bbapap.2008.06.017
14. Wang H, Vardy LA, Tan CP, et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell. 2010;18(1):52–62. doi:10.1016/j.ccr.2010.04.028
15. Peng L, Ning J, Meng L, Shou C. The association of the expression level of protein tyrosine phosphatase PRL-3 protein with liver metastasis and prognosis of patients with colorectal cancer. J Cancer Res Clin Oncol. 2004;130(9):521–526. doi:10.1007/s00432-004-0563-x
16. P W, X Y, H Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science (New York, NY). 2014;344(6181):310–313. doi:10.1126/science.1251456
17. Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6(6):4505–4515. doi:10.18632/oncotarget.2934
18. Kong L, Zhang Y, Ye ZQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–9. doi:10.1093/nar/gkm391
19. Wang L, Park HJ, Dasari S, Wang S, Kocher JP, CPAT: LW. Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):e74. doi:10.1093/nar/gkt006
20. Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27(13):i275–82. doi:10.1093/bioinformatics/btr209
21. Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi:10.1126/science.1112009
22. Halley P, Khorkova O, Wahlestedt C. Natural antisense transcripts as therapeutic targets. Drug Discov Today Ther Strateg. 2013;10(3):e119–e125. doi:10.1016/j.ddstr.2013.03.001
23. Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. 2015;16(2):3251–3266. doi:10.3390/ijms16023251
24. Guo K, Li J, Wang H, et al. PRL-3 initiates tumor angiogenesis by recruiting endothelial cells in vitro and in vivo. Cancer Res. 2006;66(19):9625–9635. doi:10.1158/0008-5472.CAN-06-0726
25. Kato H, Semba S, Miskad UA, Seo Y, Kasuga M, Yokozaki H. High expression of PRL-3 promotes cancer cell motility and liver metastasis in human colorectal cancer: a predictive molecular marker of metachronous liver and lung metastases. Clin Cancer Res. 2004;10(21):7318–7328. doi:10.1158/1078-0432.CCR-04-0485
26. Finn RS, Zhu AX, Farah W, et al. Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: a systematic review and meta-analysis. Hepatology. 2018;67(1):422–435. doi:10.1002/hep.29486
27. Lanzafame M, Bianco G, Terracciano LM, Ng CKY, Piscuoglio S. The Role of Long Non-Coding RNAs in Hepatocarcinogenesis. Int J Mol Sci. 2018;19(3):682. doi:10.3390/ijms19030682
28. DiStefano JK. Long noncoding RNAs in the initiation, progression, and metastasis of hepatocellular carcinoma. Noncoding RNA Res. 2017;2(3–4):129–136. doi:10.1016/j.ncrna.2017.11.001
29. Fang TT, Sun XJ, Chen J, et al. Long non-coding RNAs are differentially expressed in hepatocellular carcinoma cell lines with differing metastatic potential. Asian Pac J Cancer Prev. 2014;15(23):10513–10524. doi:10.7314/apjcp.2014.15.23.10513
30. Fahey ME, Moore TF, Higgins DG. Overlapping antisense transcription in the human genome. Comp Funct Genomics. 2002;3(3):244–253. doi:10.1002/cfg.173
31. Babajko S, Petit S, Fernandes I, et al. Msx1 expression regulation by its own antisense RNA: consequence on tooth development and bone regeneration. Cells Tissues Organs. 2009;189(1–4):115–121. doi:10.1159/000151748
32. Xu Z, Wei W, Gagneur J, et al. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7(1):468. doi:10.1038/msb.2011.1
33. Rintala-Maki ND, Sutherland LC. Identification and characterisation of a novel antisense non-coding RNA from the RBM5 gene locus. Gene. 2009;445(1–2):7–16. doi:10.1016/j.gene.2009.06.009
34. Wang Y, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi:10.1016/j.stem.2015.03.003
35. Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10(9):637–643. doi:10.1038/nrm2738
36. Aladesuyi Arogundade O, Stauffer JE, Saberi S, et al. Antisense RNA foci are associated with nucleoli and TDP-43 mislocalization in C9orf72-ALS/FTD: a quantitative study. Acta Neuropathol. 2019;137(3):527–530. doi:10.1007/s00401-018-01955-0
37. Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8(11):1311–1323. doi:10.1101/gad.8.11.1311
38. Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi:10.1038/nature09819
39. Castillo-Quan JI, Blackwell TK. Metformin: restraining nucleocytoplasmic shuttling to fight cancer and aging. Cell. 2016;167(7):1670–1671. doi:10.1016/j.cell.2016.11.058
40. Mayinuer A, Yasen M, Mogushi K, et al. Upregulation of protein tyrosine phosphatase type IVA member 3 (PTP4A3/PRL-3) is associated with tumor differentiation and a poor prognosis in human hepatocellular carcinoma. Ann Surg Oncol. 2013;20(1):305–317. doi:10.1245/s10434-012-2395-2
41. Zhao WB, Li Y, Liu X, Zhang LY, Wang X. Evaluation of PRL-3 expression, and its correlation with angiogenesis and invasion in hepatocellular carcinoma. Int J Mol Med. 2008;22(2):187–192.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.