Back to Journals » Cancer Management and Research » Volume 13
LncRNA PCAT18 Promotes Non-Small Cell Lung Cancer Progression by Sponging miR-4319
Authors He L, Wang J, Zhou L, Li X
Received 25 December 2020
Accepted for publication 25 March 2021
Published 10 May 2021 Volume 2021:13 Pages 3761—3774
DOI https://doi.org/10.2147/CMAR.S298918
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Li He,1 Jianjun Wang,2 Long Zhou,3 Xiaobing Li1
1Department of Oncology, The People’s Hospital of Xinyu City, Xinyu, Jiangxi, People’s Republic of China; 2Department of Radiology, Haiyan People’s Hospital, Jiaxing, Zhejiang, People’s Republic of China; 3Department of Radiation Oncology, Xiangtan Central Hospital, Xiangtan, Hunan, People’s Republic of China
Correspondence: Xiaobing Li
Department of Oncology, The People’s Hospital of Xinyu City, 369 Xinxinbei Road, Xinyu, Jiangxi, 330008, People’s Republic of China
Email [email protected]
Introduction: NSCLC (non-small cell lung cancer), the most common type of human cancer, is a main cause of cancer-associated mortality. Accumulating evidence has confirmed that long non-coding RNAs serve crucial roles in NSCLC development.
Methods: The PCAT18 expression in NSCLC tissues and cell lines were evaluated by reverse transcription-quantitative PCR. Cell Counting Kit-8 assays, colony formation study, wound healing assays and transwell invasion assays, and tumor xenograft experiments were performed to investigate the biological functions of PCAT18 in NSCLC. Luciferase reporter, RNA-binding protein immunoprecipitation (RIP) and RNA pull-down assays were further used to explore the association between PCAT18 and miR-4319.
Results: PCAT18 expression was up-regulated in NSCLC tissues and cell lines. Furthermore, PCAT18 silencing inhibited NSCLC cell proliferation, migration and invasion, while co-transfection with a miR-4319 inhibitor reversed these biological effects, and miR-4319 inhibited NSCLC growth in vivo. Additionally, PCAT18 silencing promoted NSCLC cell apoptosis and induced G1 stage arrest. Moreover, luciferase reporter assays illustrated that PCAT18 regulated miR-4319 directly, and a RIP assay and RNA pull-down analysis further demonstrated that miR-4319 inhibited PCAT18 in a RNA-induced silencing complex-dependent manner. Finally, PCAT18 silencing impaired the growth of NSCLC in vivo.
Conclusion: In conclusion, these findings demonstrated that PCAT18 promoted NSCLC development by sponging miR-4319. PCAT18 may serve as a crucial biomarker for the diagnosis and targeted therapy of NSCLC.
Keywords: PCAT18, miR-4319, non-small cell lung cancer, invasion, proliferation
Introduction
Lung cancer has been recognized as the most common type of human cancer, also the predominant reason of cancer-related mortality worldwide.1 Statistical analysis has demonstrated that NSCLC accounts for ~80% of all lung cancer cases, which include squamous cell carcinoma, adenosquamous cell carcinoma, adenocarcinoma and large cell carcinoma.2,3 Although great progression has been made in the clinical diagnosis and therapies, such as chemotherapy, surgical resection, radiotherapy, molecular targeted therapy and immunotherapy, patients with NSCLC still face an extremely poor prognosis.4 Therefore, elucidation of the molecular mechanism and identification of new potential therapeutic targets for NSCLC is imperative to improve the rather dismal prognosis of NSCLC patients.
Long non-coding RNAs (lncRNAs) are defined as transcripts with a length of >200 nucleotides.5,6 Emerging researches have suggested that lncRNAs serve crucial functions in the initiation and development of human cancer.7,8 Numerous studies have demonstrated that lncRNAs were involved in NSCLC progression.9,10
LncRNA PCAT18 was a recently identified lncRNA. It was reported that PCAT18 inhibited gastric cancer progression.11,12 Nevertheless, Yang et al reported the oncogenic role of PCAT18 in colorectal cancer.13 Obviously, the biological effects and underlying mechanism of PCAT18 in the development of main human cancers, including NSCLC, remain elusive.
The present study assessed the expression of PCAT18 in NSCLC tissues and paired adjacent normal lung tissues. Furthermore, the biological functions of PCAT18 in NSCLC were assessed in vitro and in vivo. Moreover, the current study also examined the association between PCAT18 and miR-4319 to reveal the molecular mechanism of PCAT18 in NSCLC development. To the best of our knowledge, this study was the first to reveal that PCAT18 exerts oncogenic roles in the progression of NSCLC.
Materials and Methods
Clinical Samples
Totally, 30 NSCLC tissues and paired adjacent normal lung tissues were obtained from NSCLC patients (age, 43–72 years; male patients, 17; female patients, 13) undergoing surgical resection at the People’s Hospital of Xinyu city (Xinyg, China) from May 2015 to September 2018. All included patients did not receive chemotherapy or radiation before collection of the specimens. All participants signed informed consent forms prior to sample collection. The samples collected during operation were rapidly frozen in liquid nitrogen until further use. This study was approved by the Ethic and Research Committees of the People’s Hospital of Xinyu city (Xinyu, China). This study was conducted in accordance with the Declaration of Helsinki.
Cell Culture
The normal lung HBE cell line and human NSCLC H460, H520, A549 and H1299 cell lines were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. The cells were cultured in DMEM supplemented with 10% fetal calf serum, 50 U/mL penicillin (Biowest) and 0.1 mg/mL streptomycin (Biowest). All cell cultures were maintained at 37°C in a humidified incubator with 5% CO2.
Constructs, Synthesized Oligos and Transfection
The short hairpin RNA (shRNA/sh) targeting PCAT18 (sh- PCAT18), miR-4319 mimics, inhibitors and their corresponding negative controls (NCs) were purchased from Shanghai GeneChem Co., Ltd. All DNAs were inserted into pcDNA3.1. Finally, Lipofectamine 3000 was utilized to transfer the oligonucleotides and constructs into the A549 and H1299 cells. The concentration of each miRNA transfected was 50 nM per well. At 36–48 h after transfection, the cells were used for subsequent experiments.
RNA Extraction and RT-qPCR
The total RNA was obtained from tissues and cells adopting TRIzol reagent. A total of 2 µg RNA was reverse transcribed into cDNA adopting the PrimeScript RT reagent kit. RT-qPCR was conducted using SYBR Green Master Mix on an ABI PRISM 7500 PCR system. GAPDH and U6 were used as controls and for the normalization of the expression levels of mRNA and miRNA, respectively. The primer sequences of RNAs are shown in Table 1.
 |
Table 1 Primer List |
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8) and colony formation assays were performed to evaluate the proliferation ability. For the CCK-8 assay, cells were seeded in 96-well plates (3000 cells/well), then 10 μL CCK-8 solution was added after 24, 48, 72 and 96 h of culture. After 2 h, the plates were washed using PBS and finally the absorbance at 450 nm was measured using a microplate reader. For colony formation analysis, 1000 cells were seeded into a 6-well plate and continuously incubated for 12 days. The colonies were fixed using 4% paraformaldehyde and stained with 1% crystal violet. Finally, the colonies were counted and imaged.
Wound Healing Assay
A sterile pipette tip (p200) was adopted to make a linear wound when the cells had grown to nearly 100% confluence. Subsequently, the cells were washed 3 times using PBS and then cultured with DMEM without serum at 37°C for 24 h. Finally the images were obtained at 0 and 24 h after scratching using an inverted microscope (magnification, x200; Nikon Corporation).
Transwell Invasion Assay
The transwell invasion assays were conducted to determine the cell invasion potential using transwell plates coated with 50 µL Matrigel (BD Biosciences). Briefly, 1×105 cells were suspended in 300 μL serum-free medium and added to the upper chamber, while 800 μL complete medium was placed in the lower chamber. 24 h later, cells on the upper surface of the membrane were scraped off, while the cells on the lower side of the chamber were fixed and stained. The invaded cells were counted in more than 5 fields under a light microscope.
Luciferase Reporter Assay
Mut (mutant-type) or wt (wild-type) fragments of PCAT18 containing the miR-4319 targeting site were synthesized and cloned into a dual-luciferase reporter vector (pmirGLO; Shanghai GenePharma Co., Ltd.). Similarly, luciferase vectors and miR-4319 mimics or miR-4319 NC and Renilla plasmid were co-transfected into A549 cells using Lipofectamine 3000. 48 hours after transfection, a dual-luciferase assay was used to examine the Renilla and firefly luciferase activity according to the manufacturer’s protocol, and the levels of firefly luciferase activity were normalized to that of Renilla luciferase activity.
RNA-Binding Protein Immunoprecipitation (RIP) Assay
A Magna RIP RNA-Binding Protein Immunoprecipitation Kit (EMD Millipore) was adopted for the RIP assay. Cells were harvested and lysed. Lysis buffer containing magnetic beads was incubated with human Ago2 (anti-argonaute RISC catalytic component 2) antibody (cat. no. ab32381; 1:1000; Abcam) to conjugate the antibody to the magnetic beads. Subsequently, proteinase K was added to digest the protein, and the immunoprecipitated RNAs were isolated using TRIzol reagent and measured.
RNA Pull-Down Assay
A549 cells were transfected with biotin-labeled miR-4319 mimic or NC. After 24 h, cells were collected and cultured with M-280 streptavidin magnetic beads (Invitrogen; Thermo Fisher Scientific, Inc.) at 4°C for 4 h with rotation. Subsequently, the beads were washed using lysis buffer containing proteinase K and 10% SDS, then the supernatants were obtained and the RNA was isolated and coprecipitated RNA was detected using RT-qPCR assays.
Tumor Xenograft Experiment
Ten female BALB/c nude mice (6–8 weeks old, 18–22 g) were bought from the Animal Center of of Nanchang University. H1299 cells (2x106) that were stably transfected with lv-sh-PCAT18 or lv-sh-NC were injected subcutaneously into the left flank of the mice (n=5 mice per group). Tumor sizes were measured every week. After 4 weeks, the mice were anesthetized and the tumor tissues were collected and weighed, and RT-qPCR was conducted to examine the expression of PCAT18 and miR-4319. This study was approved by the Ethic and Research Committees of the People’s Hospital of Xinyu city (Xinyg, China). The guidelines for the welfare of experimental animals are GB/T 35892–2018 standard issued by the General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China.
Statistical Analysis
The experiments were repeated at least three times. All results are presented as the mean ± SD. All statistical analyses were conducted using GraphPad Prism software. One-way ANOVA with Bonferroni post hoc test or two-tailed Student’s t-test were used for the comparisons among groups. In addition, Pearson’s coefficient correlation analysis was performed for expression correlation analysis. P<0.05 was recognized to indicate a statistically significant difference.
Results
PCAT18 Expression is Elevated in NSCLC
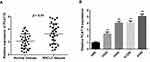
RT-qPCR was conducted to assess the relative expression levels of PCAT18 in 30 pairs of NSCLC and adjacent normal tissues. PCAT18 expression was markedly increased in NSCLC tissues compared with in paired normal tissues (Figure 1A). Moreover, the expression levels of PCAT18 in four NSCLC cell lines and in the normal human lung HBE cell line were examined. The results indicated that the NSCLC cell lines exhibited markedly higher expression levels of PCAT18 compared with the normal lung HBE cell line (Figure 1B).
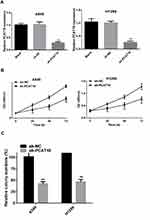 |
Figure 2 Continued. |
PCAT18 Silencing Suppresses NSCLC Cell Proliferation, Migration and Invasion
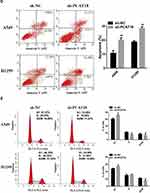
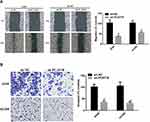
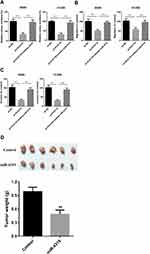
In order to further explore the function of PCAT18 in NSCLC, sh-PCAT18 was transfected into A549 and H1299 cells, and RT-qPCR was conducted to confirm the transfection efficiency (Figure 2A). Next, CCK-8 assays and colony formation assays were conducted to explore the effects of PCAT18 on NSCLC cell proliferation, migration and invasion. The results suggested that the proliferation of the A549 and H1299 cells in the sh-PCAT18 group was markedly impaired compared with that of cells in the NC group (Figure 2B and C). Moreover, knockdown of PCAT18 increased apoptosis (Figure 2D) and induced cell cycle arrest in the G1 phase (Figure 2E) of A540 and H1299 cells. Consistently, the results of wound healing and transwell invasion assays suggested that sh-PCAT18 markedly suppressed NSCLC cell migration and invasion (Figure 3A and B). Overall, PCAT18 silencing impaired NSCLC cell proliferation, migration and invasion in vitro.
Reciprocal Modulation Between PCAT18 and miR-4319)
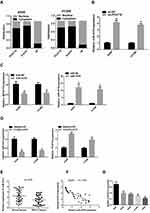
To investigate the potential mechanism of PCAT18 in the development of NSCLC, we determined the subcellular localization of PCAT18. The results showed that PCAT18 was mostly distributed in the cytoplasm (Figure 4A), which suggested that PCAT18 might exert its biological function by sponging miRNA. miRcode (http://www.mircode.org) was used to conduct bioinformatics analysis, and the results indicated that miR-4319 was a potential target of PCAT18. To confirm this hypothesis, the correlation between PCAT18 and miR-4319 was explored. The present results demonstrated that miR-4319 expression was markedly increased following knockdown of PCAT18 (Figure 4B). Furthermore, PCAT18 expression was markedly decreased when the cells were transfected with miR-4319 mimic (Figure 4C), while the expression levels of PCAT18 were markedly increased when the cells were transfected with miR-4319 inhibitor (Figure 4D). In addition, miR-4319 expression was determined in NSCLC tissues, and the results indicated that miR-4319 expression was down-regulated in NSCLC tissues (Figure 4E). Notably, miR-43199 expression was negatively correlated with PCAT18 expression (Figure 4F). Additionally, NSCLC cell lines exhibited markedly lower expression levels of miR-4319 compared with the normal lung HBE cell line (Figure 4G).
miR-4319 is a Direct Target of PCAT18
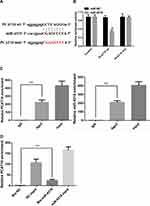
A dual-luciferase reporter assay was conducted to further assess whether miR-4319 was a direct target of PCAT18. The data of the luciferase assay revealed that miR-4319 mimic markedly decreased luciferase reporter expression in the cells transfected with PCAT18-wt but not in the cells transfected with PCAT18-mut or NC (Figure 5A and B). It is widely acknowledged that miRNAs function by regulating RNA-induced silencing complex (RISC).14 Ago2, a key component of RISC, exerts crucial roles in RNA cleavage. Therefore, a RIP assay was conducted to determine whether miR-4319 regulated PCAT18 via RISC formation. As the results demonstrated, compared with NC (IgG), PCAT18 and miR-4319 were preferentially enriched in anti-Ago2 antibody-incubated beads (Figure 5C). To further investigate whether PCAT18 and miR-4319 bind to each other, RNA pull-down assay was also performed. The present results demonstrated that compared with NC, the biotin-labeled miR-4319 mimic pulled down more PCAT18 (Figure 5D). Collectively, these data revealed that PCAT18 bound to miR-4319 directly.
Biological Function of PCAT18 in NSCLC Cells is Dependent on miR-4319
Rescue experiments were performed to investigate whether PCAT18affected NSCLC cell proliferation, migration and invasion in a miR-4319-dependent manner. The colony formation assays demonstrated that PCAT18 silencing-induced impairment of proliferation in NSCLC cells was partially abolished in the presence of miR-4319 inhibitor (Figure 6A). Additionally, miR-4319 inhibitor reversed the PCAT18 silencing-induced inhibitory effect on migration and invasion in NSCLC cells (Figure 6B and C). Moreover, miR-4319 inhibited NSCLC growth in vivo (Figure 6D). All these results demonstrated that the oncogenic function of PCAT18 in NSCLC involved negative regulation of miR-4319.
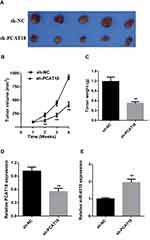
PCAT18 Silencing Suppresses NSCLC Tumor Growth in vivo
To further verify the in vitro results, a subcutaneous xenograft tumor model was established by injecting stable sh-NC or sh-PCAT18 cells into nude mice. Consistent with the in vitro findings, the in vivo experiments indicated that the volume and weight of tumors in the sh- PCAT18 group were markedly reduced compared with those in the sh-NC group (Figure 7A–C). Furthermore, the expression levels of PCAT18 were decreased in the sh-PCAT18 group tumors, while miR-4319 expression was increased in the sh- PCAT18 group tumors compared with in the sh-NC group tumors (Figure 7D and E). Therefore, it was concluded that PCAT18 silencing suppressed NSCLC tumor growth in vivo.
Discussion
In recent years, accumulating evidence has revealed fundamental roles of lncRNAs in the regulation of numerous physiological and pathological processes, such as cell differentiation, proliferation and apoptosis, and the initiation and progression of human cancer.7,15,16 At present, it is widely acknowledged that numerous lncRNAs are involved the progression of NSCLC. For example, long noncoding RNA KCNMB2-AS1 increased ROCK1 expression by Sponging microRNA-374a-3p to facilitate the progression of NSCLC.17 LncRNA F630028O10Rik, a novel tumour suppressor, inhibited lung cancer angiogenesis by regulating miR-223-3p.18 MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in NSCLC cells.19 However, the exact biological function and mechanism of the majority of lncRNAs in NSCLC remain unclear.
PCAT18 (prostate cancer-associated transcript 18) was first found that, in metastatic prostate cancer, PCAT18 silencing inhibited cell migration and invasion.20 Moreover, it was reported that PCAT18 was down-regulated in gastric cancer and inhibited gastric cancer progression through miR-107/PTEN/PI3K/AKT signaling pathway.11 Besides, Zhang et al reported that PCAT18 suppressed gastric cancer cells proliferation, migration and invasion via miR-135b inhibition to increase CLDN11 expression.12 Nevertheless, Yang et al found that PCAT18 was highly expressed in colorectal cancer tissues and cells, and the expression of PCAT18 was positively associated with progression of colorectal cancer patients. Furthermore, PCAT18 promoted colorectal cancer cell proliferation, migration and invasion through binding miR-759 to promote the expression of SPRR.13 The biological roles and mechanism of PCAT18 in human cancers are largely unknown. The current study aimed to investigate the exact roles and potential mechanism of PCAT18 in NSCLC. First, it was observed that the PCAT18 expression was up-regulated in clinical NSCLC tissues and cell lines. Second, our study also evaluated the biological effects of PCAT18 silencing on NSCLC cell proliferation, migration and invasion in vitro, and tumor growth in vivo. The results of a series of experiments revealed that PCAT18 silencing inhibited cell proliferation, migration and invasion in vitro and tumor growth in vivo in NSCLC.
The competing endogenous RNA (ceRNA) theory, wherein miRNA is sequestered by lncRNA which serves as a molecular sponge, has attracted increasing attention.21,22 For instance, lncRNA POU6F2-AS2 drives the development and drug resistance of colon cancer via the miR-377/BRD4 axis.23 Long non-coding RNA Linc00485 promoted lung cancer development by regulating miR-298/c-Myc axis.24 LncRNA SNHG15 modulated EGFR-TKI acquired resistance in lung adenocarcinoma by sponging miR-451 to up-regulate MDR-1 expression.25 Therefore, it was predicted that miR-4319 may be a direct target of PCAT18. In this study, PCAT18 was revealed to be a ceRNA by directly targeting miR-4319. Previous studies have demonstrated that miR-4319 involved the development of several types of human cancer. For instance, miR-4319 inhibited of epithelial-mesenchymal transition and impaired stemness of HCC cells through targeting FOXQ1.26 KCNQ1OT1 accelerates gastric cancer progression via miR-4319/DRAM2 axis.27 MiR-4319 inhibited esophageal squamous cell carcinoma proliferation through regulating NLRC5.28 The results of the RT-qPCR indicated the reciprocal inhibitory effect of PCAT18 and miR-4319. Furthermore, the effect of PCAT18 silencing on NSCLC cell proliferation, migration and invasion was reversed by miR-4319 inhibitor. The results of the luciferase assay showed that miR-4319 directly bound to PCAT18. Additionally, the data of RIP and RNA pull-down assays revealed that miR-4319 modulated PCAT18 in a RISC-dependent manner. Collectively, these data demonstrated that PCAT18 acts as an endogenous sponge of miR-4319, and that PCAT18 and miR-4319 negatively regulate each other in NSCLC.
Disclosure
The authors reported no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Yu C, Cheng Z, Cui S, et al. circFOXM1 promotes proliferation of non-small cell lung carcinoma cells by acting as a ceRNA to upregulate FAM83D. J Exp Clin Cancer Res. 2020;39(1):55. doi:10.1186/s13046-020-01555-5
3. He H, Song X, Yang Z, et al. Upregulation of KCNQ1OT1 promotes resistance to stereotactic body radiotherapy in lung adenocarcinoma by inducing ATG5/ATG12-mediated autophagy via miR-372-3p. Cell Death Dis. 2020;11(10):883. doi:10.1038/s41419-020-03083-8
4. Mustachio LM, Roszik J, Farria AT, Guerra K, Dent SY. Repression of GCN5 expression or activity attenuates c-MYC expression in non-small cell lung cancer. Am J Cancer Res. 2019;9(8):1830–1845.
5. Song Z, Zhang X, Lin Y, Wei Y, Liang S, Dong C. LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J Cell Mol Med. 2019;23(11):7554–7565. doi:10.1111/jcmm.14625
6. Han Q, Li J, Xiong J, Song Z. Long noncoding RNA LINC00514 accelerates pancreatic cancer progression by acting as a ceRNA of miR-28-5p to upregulate Rap1b expression. J Exp Clin Cancer Res. 2020;39(1):151. doi:10.1186/s13046-020-01660-5
7. Wang Z, Cheng Y, Zhu Y, et al. Long non-coding RNA FOXD1-AS1 promotes the progression and glycolysis of nasopharyngeal carcinoma by sustaining FOXD1 expression. Am J Cancer Res. 2020;10(11):3686–3704.
8. Meng L, Xing Z, Guo Z, Liu Z. LINC01106 post-transcriptionally regulates ELK3 and HOXD8 to promote bladder cancer progression. Cell Death Dis. 2020;11(12):1063. doi:10.1038/s41419-020-03236-9
9. Guan Y, Yang J, Liu X, Chu L. Long noncoding RNA CBR3 antisense RNA 1 promotes the aggressive phenotypes of nonsmallcell lung cancer by sponging microRNA5093p and competitively upregulating HDAC9 expression. Oncol Rep. 2020;44(4):1403–1414. doi:10.3892/or.2020.7719
10. Dai J, Wang B, Zhao Y, et al. Long noncoding RNA LINC01426 sequesters microRNA-519d-5p to promote non-small cell lung cancer progression by increasing ETS1 expression. Cancer Manag Res. 2020;12:12697–12708. doi:10.2147/CMAR.S277113
11. Chen P, Zhao X, Wang H, Zheng M, Wang Q, Chang W. The down-regulation of lncRNA PCAT18 promotes the progression of gastric cancer via MiR-107/PTEN/PI3K/AKT signaling pathway. Onco Targets Ther. 2019;12:11017–11031. doi:10.2147/OTT.S225235
12. Zhang XZ, Mao HL, Zhang SJ, et al. lncRNA PCAT18 inhibits proliferation, migration and invasion of gastric cancer cells through miR-135b suppression to promote CLDN11 expression. Life Sci. 2020;249:117478. doi:10.1016/j.lfs.2020.117478
13. Yang D, Li R, Xia J, et al. Long noncoding RNA PCAT18 upregulates SPRR3 to promote colorectal cancer progression by binding to miR-759. Cancer Manag Res. 2020;12:11445–11452. doi:10.2147/CMAR.S272652
14. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi:10.1038/nrg2290
15. Liu Y, Chen X, Chen X, et al. Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis. 2020;11(3):175. doi:10.1038/s41419-020-2268-8
16. Chen L, Ren P, Zhang Y, Gong B, Yu D, Sun X. Long noncoding RNA GAS5 increases the radiosensitivity of A549 cells through interaction with the miR21/PTEN/Akt axis. Oncol Rep. 2020;43(3):897–907. doi:10.3892/or.2020.7467
17. Yang H, Wang Z, Wang Z. Long noncoding RNA KCNMB2-AS1 increases ROCK1 expression by sponging microRNA-374a-3p to facilitate the progression of non-small-cell lung cancer. Cancer Manag Res. 2020;12:12679–12695. doi:10.2147/CMAR.S270646
18. Qin L, Zhong M, Adah D, et al. A novel tumour suppressor lncRNA F630028O10Rik inhibits lung cancer angiogenesis by regulating miR-223-3p. J Cell Mol Med. 2020;24(6):3549–3559. doi:10.1111/jcmm.15044
19. Gai C, Liu C, Wu X, et al. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis. 2020;11(9):751. doi:10.1038/s41419-020-02939-3
20. Crea F, Watahiki A, Quagliata L, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. doi:10.18632/oncotarget.1769
21. Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi:10.1016/j.cell.2011.09.028
22. Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, Stoffel M. Impact of microRNA levels, target-site Complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol Cell. 2016;64(3):565–579. doi:10.1016/j.molcel.2016.09.027
23. Xu G, Zhu H, Xu J, et al. Long non-coding RNA POU6F2-AS2 promotes cell proliferation and drug resistance in colon cancer by regulating miR-377/BRD4. J Cell Mol Med. 2020;24(7):4136–4149. doi:10.1111/jcmm.15070
24. Zhang Z, Lin W, Lin Y, et al. Long intergenic non-coding RNA Linc00485 promotes lung cancer progression by modulating miR-298/c-Myc axis. J Cell Mol Med. 2020.
25. Huang J, Pan B, Xia G, Zhu J, Li C, Feng J. LncRNA SNHG15 regulates EGFR-TKI acquired resistance in lung adenocarcinoma through sponging miR-451 to upregulate MDR-1. Cell Death Dis. 2020;11(7):525. doi:10.1038/s41419-020-2683-x
26. Han S, Shi Y, Sun L, Liu Z, Song T, Liu Q. MiR-4319 induced an inhibition of epithelial-mesenchymal transition and prevented cancer stemness of HCC through targeting FOXQ1. Int J Biol Sci. 2019;15(13):2936–2947. doi:10.7150/ijbs.38000
27. Wang J, Wu F, Li Y, et al. KCNQ1OT1 accelerates gastric cancer progression via miR-4319/DRAM2 axis. Int J Immunopathol Pharmacol. 2020;34:2058738420954598. doi:10.1177/2058738420954598
28. Hu X, Wang M, Cao L, et al. miR-4319 suppresses the growth of esophageal squamous cell carcinoma via targeting NLRC5. Curr Mol Pharmacol. 2020;13(2):144–149. doi:10.2174/1874467212666191119094636
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.