Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA OR3A4 Regulated the Growth of Osteosarcoma Cells by Modulating the miR-1207-5p/G6PD Signaling
Authors Wang X, Chen K, Zhao Z
Received 13 October 2019
Accepted for publication 20 January 2020
Published 15 April 2020 Volume 2020:13 Pages 3117—3128
DOI https://doi.org/10.2147/OTT.S234514
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Xiaole Wang, Kunfeng Chen, Zhijian Zhao
Department of Traumatology, The First People’s Hospital of Shangqiu, Shangqiu, Henan Province 476000, People’s Republic of China
Correspondence: Xiaole Wang
Department of Traumatology, The First People’s Hospital of Shangqiu, No. 292 Kaixuan South Road, Suiyang District, Shangqiu City, Henan Province 476000 Email [email protected]
Background: Increasing evidence has demonstrated the importance of non-coding RNAs including long non-coding RNA (lncRNA) and microRNAs (miRNAs) in the tumorigenesis of osteosarcoma (OS). Abnormal expression of lncRNA olfactory receptor family 3 subfamily A member 4 (OR3A4) was found in multiple human cancers; however, the function of OR3A4 in OS remains largely unknown.
Materials and Methods: The expression level of OR3A4 in OS tissues and cell lines was detected by RT-qPCR. Cell counting kit-8 assay, colony formation and flow cytometry analysis were performed to determine the growth of OS cells. The targets of OR3A4 were predicted using the miRDB database. The binding between OR3A4 and miRNAs was confirmed by dual-luciferase reporter assay.
Results: OR3A4 was overexpressed in OS tissues and correlated with the advanced progression of OS patients. Down-regulation of OR3A4 significantly inhibited the proliferation and colony formation of OS cells. Mechanistically, OR3A4 acted as a sponge of miR-1207-5p. Glucose-6-phosphate dehydrogenase (G6PD) was identified as a target of miR-1207-5p. Knockdown of OR3A4 increased the expression of miR-1207-5p and consequently, suppressed the level of G6PD in OS cells. Due to the essential role of G6PD in the pentose phosphate pathway (PPP), depletion of OR3A4 inhibited NADPH production, glucose consumption and lactate generation. Decreased level of NADPH by depletion of OR3A4 up-regulated the redox state (ROS) content and resulted in endoplasmic reticulum (ER) stress in OS cells. Restoration of G6PD significantly attenuated the cell growth inhibition induced by OR3A4 knockdown.
Conclusion: Our finding suggested the critical role of OR3A4 in the proliferation of OS cells via targeting the miR-1207-5p/G6PD axis.
Keywords: osteosarcoma, OR3A4, miR-1207-5p, G6PD, NADPH
Introduction
Osteosarcoma (OS) is one of the most common cancers and the leading cause of cancer-related death around the world, which is usually diagnosed at an advantaged metastatic stage.1,2 Currently, the first-line therapy of patient with OS includes surgery resection, cisplatin-based chemotherapy and radiotherapy.3 Due to the progression in the clinical practice, the 5-year survival rate of OS patients has been improved in recent decades, however, most patients eventually advance to recurrent, progressive stage due to the resistance. Therefore, exploring the mechanisms that are responsible for the development of OS is quite necessary.
An increasing body of evidence has demonstrated that long non-coding RNAs (lncRNAs, >200 nucleotides) play pivotal roles in the development of numerous malignant tumors.4,5 Whole-genome sequence analysis uncovered that thousands of lncRNAs were aberrantly expressed in cancers and regulated the proliferation, differentiation and migration of cancer cells.6–8 For another big group of non-coding RNAs, microRNAs (miRNAs) are defined as a class of small, single-stranded RNAs without the capacity of producing protein.9–11 MiRNAs act as negative regulators of gene expression via binding to the 3ʹ-untranslated region (UTR) of target mRNAs, which leads to mRNA degradation or translation inhibition.11,12 The involvement of miRNAs in the progression of OS has been demonstrated by increasing studies.13–17 Notably, recent findings showed that lncRNAs regulated gene expression by sponging miRNAs, which was also known as competing endogenous RNAs (ceRNAs).18–21 Based on this hypothesis, lncRNAs bound miRNAs and modulated the expression of miRNA-targeted genes. For example, lncRNA Taurine upregulated gene 1 promoted the metastasis of OS cells by regulating HIF-1α via sponging miR-143-5p.22 LncRNA ROR enhanced the progression of OS by targeting miR-206.23 The oncogenic function of lncRNA OR3A4 in the development of cancers has been reported recently.24–29 OR3A4 promoted the metastasis of gastric cancer and ovarian cancer.26,29 Similarly, OR3A4 enhanced the proliferation and migration of breast cancer cells.27 These findings indicated the vital roles of OR3A4 in promoting the malignant behaviors of cancer cells. However, the mechanism of OR3A4 in OS has not been characterized.
Reprogramming of glucose metabolism is one of the key characteristic features of cancer to support the rapid proliferation of cancer cells. Glucose-6-phosphate dehydrogenase (G6PD) is the rate-limiting enzyme that converts glucose metabolism from glycolysis into the pentose phosphate pathway (PPP), which is vital for the production of reduced nicotinamide adenine dinucleotide phosphate (NADPH).30 NADPH is required for the biosynthesis of lipids. Increasing evidence suggested that G6PD was up-regulated in a variety of cancers.31–33 Overexpression of G6PD was associated with the poor prognosis of cancer patients, suggesting G6PD as a promising target to interrupt the progression of cancers.
In this study, we found that OR3A4 was overexpressed in OS tissues and correlated with the advanced progression of OS patients. Down-regulation of OR3A4 inhibited the growth of OS cells. Mechanism study revealed that depletion of OR3A4 decreased G6PD via sponging miR-1207-5p, leading to the decreased NADPH production. Our results demonstrated the novel functional mechanism of the OR3A4/miR-1207-5p/G6PD axis in OS.
Materials and Methods
Tissue Samples
Fifty paired OS tissues and adjacent normal tissues were obtained from OS patients undergone resection surgery at the Frist People’s Hospital of Shangqiu from May 2011 to September 2012. Participants were not treated with chemotherapy, radiotherapy, or radiotherapy before collecting the tissues. Tissues were immediately frozen in liquid nitrogen. Written informed consents were received from all the patients. The experiments were approved by the Ethics Committee of the Frist People’s Hospital of Shangqiu.
Cell Culture
Human OS cell lines MG-63, SaoS-2, SJSA-1 and G-292 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, Sigma-Aldrich, St Louis, MO, USA) with 1% penicillin and streptomycin (Hyclone, South Logan, UT, USA). Human osteoblast HOb cells (Sigma Aldrich, St. Louis, MO, USA) were cultured with DMEM/F-12 medium (Invitrogen, Carlsbad, CA, USA). Cells were maintained at 37°C in a humidified 5% CO2 atmosphere. The shRNA-control or shRNA-OR3A4 was transiently transfected into MG-63 or SaoS-2 cells with the same final concentration using Lipofectamine 2000 regent (Invitrogen, Carlsbad, CA, USA). The transfection of miR-1207-5p mimic or control miRNA, as well as HA-vector or HA-G6PD was also performed with the same method.
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration of RNA was determined using the NanoDrop 2000 (Thermo Fisher Scientific In., USA). Reverse transcription reaction was performed using the One-Step Prime Script miRNA cDNA synthesis kit (Takara, Dalian, China) according to the protocol. The quantitative PCR of OR3A4 was performed with SYBR Green Master Mix (Invitrogen, Carlsbad, CA, USA) using the Applied Biosystems 7500 Real-time RT-PCR system (Applied Biosystems, Foster City, CA, USA). The primers were synthesized by GenePharma (Shanghai, China) and the sequences were as follows: OR3A4 forward, 5′-CCTATCCCTTTCTCTAAGA-3′ and reverse, 5′-ACTTCTGCAAAAACGTGCTG-3′; GAPDH forward, 5′-CCACATCGCTCAGACACCAT-3′ and reverse, 5′-ACCAGGCGCCCAATACG-3′; miR-1207-5p forward, 5′-GTTAGGGCAGGTGGGATG-3′ and reverse, 5′- TGTATGCGGCTGGTAAGTAG-3′; U6 RNA forward, 5′-TTATGGGTCCTAGCCTGAC-3′ and reverse, 5′-CACTATTGCGGGTCTGC-3′; G6PD forward, 5′-AACATCGCCTGCGTTATCCTC-3′ and reverse, 5′-ACGTCCCGGATGATCCCAA-3′. Data was analyzed by comparative cycle threshold (CT) method. The experiment was performed in triplicates. The original RT-qPCR data from cell lines were provided in Supplementary Table 1.
Cell Proliferation Assay
The proliferation of OS cells expressing shRNA-OR3A4 or shRNA-control was determined with the Cell Counting Kit-8 (CCK-8) assay according to the manufacturer’s instructions. Briefly, cells were seeded in the 96-well plate in triplicates with the density of 2000 cells per well., CCK-8 solution was added into each well at 1-, 2-, 3-, 4- and 5 days after transfection and incubated for 4 h at 37°C. The absorbance was measured at 450 nm wavelength using the microplate reader (Bio-Rad, Hercules, CA, USA).
Cell Apoptosis Analysis
Cells transfected with shRNA-OR3A4 or shRNA-control were cultured in 60-mm dishes. After cultured for 48 h, cells were harvested and washed twice with PBS. The cell apoptosis was determined by utilizing the Annexin V-FITC and propidium iodide (PI) double staining (Invitrogen, Carlsbad, CA, USA). Cells were resuspended with the binding buffer, and then stained with Annexin V-FITC and PI for 15 min in the dark at room temperature (RT). The cell apoptosis rate was measured by the flow cytometry (BD Biosciences) with the FL-1 (FITC) and FL-2 (PI) channels.
Western Blot
Cells were lysed with RIPA buffer (Beyotime, Shanghai, China) and the protein concentration was determined with the BCA Protein Assay Kit (Pierce, USA) according to the manufacturer’s instructions. Equal amount of protein (20 μg) protein was separated by SDS-PAGE and transferred onto PVDF membranes (Millipore, USA). After blocking with 5% non-fat milk, the membrane was probed with primary antibodies overnight at 4°C. The membrane was then incubated with the HRP-conjugated secondary antibody for 1 h at RT. The expression of GAPDH was detected as the loading control. The protein bands were visualized using the ECL detection kit (Pierce, USA). Data were analyzed using the ImageQuant LAS 4000min Image system (GE, Fairfield, USA). Antibodies including G6PD (#8866, Cell Signaling Technology, Danvers, MA, USA), PERK (#5683; Cell Signaling Technology, Danvers, MA, USA), phosphor-IRE1 (Ser724) (#ab48187; Abcam, Cambridge, UK), caspase-3 (#ab184787, Abcam, Cambridge, UK), PARP (#9542; Cell Signaling Technology, Danvers, MA, USA) and GAPDH (#5174, Cell Signaling Technology, Danvers, MA, USA) were commercially obtained.
Bioinformatics Analysis
The potential binding between OR3A4 and miR-1207-5p, miR-1207-5p and the 3ʹ-UTR of G6PD were predicted with the miRDB database (http://mirdb.org/custom.html).
Luciferase Reporter Assay
The wild-type or mutated OR3A4 sequences containing the putative binding sites of miR-1207-5p were amplified and constructed into the luciferase reporter vector psi-CHECK-2 (Promega, Madison, WI, USA). Cells were co-transfected with luciferase vectors and miR-1207-5p mimics or miR-Control. After transfection for 48 h, cells were collected and the luciferase activity was determined with the Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Renilla luciferase activity was detected for the normalization.
Glucose Consumption Assay
The glucose consumption of OS cells was measured using the Glucose Uptake Assay Kit (Colorimetric, Ab136955, Abcam, Cambridge, UK) according to the manufacturer’s instruction. OS cells expressing shRNA-control or shRNA-OR3A4 were cultured overnight in medium without serum. After that, cells were washed twice with pre-cold PBS and incubated with KRPH/2% BSA for 30 mins followed by incubating with 2-deoxyglucose (2-DG) for 20 mins. Thirty microlitres of standard solution of 2-DG6P, supernatant of shRNA-control and shRNA-OR3A4 was set up in triplicates, respectively. The mix A buffer containing 8 μL of assay buffer and 2 μL of enzyme mix was added into each reaction and incubated for 1 h. And then, 90 μL of extraction buffer was added and heated at 90ºC for 40 mins. Thirty-eight microlitres of mix B buffer was added. The absorbance of each well was measured at the wavelength of 412 nm with the microplate reader at 37ºC in the dark. Normalization was performed by measuring the protein concentration using the BCA kit (Beyotime, Shanghai, China).
Determination of Lactate Production
The lactate production of OS cells expressing control-shRNA or shRNA-OR3A4 was detected using the Lactate Assay Kit (#MAK064, Sigma-Aldrich, USA). After transfection for 48 h, cells were harvested and homogenized with 100 μL of lactate assay buffer. After centrifuged at 12,000 g for 5 mins, the supernatant was deproteinized using the spin filter (10 kDa MWCO). Fifty microlitres of supernatant was added into the 96-well plate. To measure the content of lactate, the standard curve was made by adding 0, 2, 4, 6, 8 and 10 μL of the standard solution into the 96-well plate. Fifty microlitres of lactate assay buffer and master reaction mix was added and incubated for 30 mins at RT. The absorbance of each well at 570 nm was measured with the microplate reader. Normalization was performed by measuring the protein concentration with the BCA kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions.
Detection of Intracellular ROS
OS cells were transfected with shRNA-OR3A4 or co-transfected with G6PD for 36 h. The intracellular ROS level was measured using the Cellular Reactive Oxygen Species Detection Assay Kit (Abcam, USA) according to the manufacturer’s instructions.
Statistical Analysis
Data were presented as mean ± standard deviation (SD). Data analysis was performed using SPSS 19.0. Student’s t test and one-way analysis of variance (ANOVA) were applied to determine the significance between groups. P<0.05 was considered as statistically significant.
Results
OR3A4 Was Up-Regulated in OS Tissues and Cell Lines
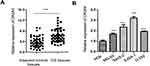
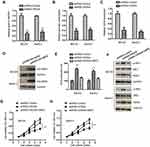
To evaluate the potential involvement of OR3A4 in OS, the expression pattern of OR3A4 in paired OS and adjacent normal tissues was detected by RT-qPCR. The result showed that OR3A4 was significantly up-regulated in OS tissues compared with that of the adjacent normal tissues (Figure 1A). Additionally, the levels of OR3A4 in cancer cell lines including MG-63, SaoS-2, SJSA-1 and G-292 were also evaluated by RT-qPCR. As indicated in Figure 1B, increased abundance of OR3A4 was observed in OS cell lines compared with that of the normal Hob cells. These results suggested the overexpression of OR3A4 in OS. To further characterize the potential clinical significance of OR3A4 in OS, the relationship between the expression of OR3A4 with the clinical features of OS patients was analyzed. The results showed that a higher level of OR3A4 was significantly correlated with the tumor size, lymph node metastasis and higher TNM stage of patients with OS (Table 1). These findings indicated the potential critical role of OR3A4 in the development of OS.
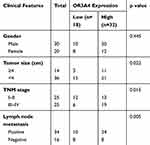 |
Table 1 Correlation Between OR3A4 Expression and Clinical Features of OS Patients |
Down-Regulation of OR3A4 Inhibited the Proliferation and Induced Apoptosis of OS
As OR3A4 was up-regulated in OS, to investigate the function of OR3A4 in OS, OR3A4 was down-regulated in OS cells by transfecting shRNA-OR3A4 into both MG-63 and SaoS-2 cells. The knockdown efficiency of OR3A4 was confirmed by RT-qPCR (Figure 2A). The effect of down-regulated OR3A4 on the proliferation of OS cells was determined by CCK-8 assay. Depleted OR3A4 markedly inhibited the proliferation of both MG-63 and SaoS-2 cells (Figure 2B and C). The inhibitory effect of OR3A4 depletion on the growth of OS cells was further evaluated by colony formation assay. As indicated in Figure 2D, the colony-forming capacity of OS cells was significantly reduced with the down-regulation of OR3A4. Meanwhile, the apoptosis percentage of OS cells harboring shRNA-OR3A4 or shRNA-control was also determined by the flow cytometry. The data showed that depletion of OR3A4 obviously enhanced the apoptosis of both MG-63 and SaoS-2 cells (Figure 2E). Consistent with the increased apoptosis, down-regulation of OR3A4 also promoted the cleavage of caspase-3 and PARP, which are the markers of cell apoptosis (Figure 2F). Collectively, these findings suggested that down-regulation of OR3A4 suppressed the growth of OS cells.
OR3A4 Sponged miR-1207-5p in OS Cells
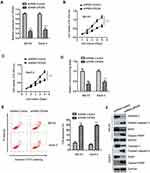
Increasing evidence has demonstrated that lncRNAs acted as competing endogenous RNAs to sponge the function of miRNAs, thereby relieving the repression of target mRNAs. To further characterize the molecular mechanism of OR3A4 in regulating the malignant behaviors of OS, the potential binding between OR3A4 and miRNAs was predicted with the bioinformatics analysis. The putative binding site of OR3A4 within the sequence of miR-1207-5p was found (Figure 3A). To investigate the function of miR-1207-5p, miR-1207-5p was overexpressed by transfecting miR-1207-5p mimics into OS cells (Figure 3B). To confirm the prediction, luciferase reporter assay was performed by co-transfecting wild-type (WT) or mutant (Mut) OR3A4 and miR-1207-5p mimics into OS cells. The results showed that overexpression of miR-1207-5p significantly inhibited the luciferase activity of WT but not the Mut OR3A4 (Figure 3C and D). To evaluate whether the interaction between OR3A4 and miR-1207-5p affected the stability of miR-1207-5p, the expression of miR-1207-5p in OS cells with depleted OR3A4 was detected by RT-qPCR. As indicated in Figure 3E, down-regulation of OR3A4 markedly increased the expression of miR-1207-5p in both MG-63 and SaoS-2 cells.
As OR3A4 was up-regulated in OS and OR3A4 negatively modulated the expression of miR-1207-5p, the expression pattern of miR-1207-5p in paired OS tissues and adjacent normal tissues was detected by RT-qPCR analysis. Results showed that miR-1207-5p was significantly decreased in OS tissues compared with that of the non-cancer tissues (Figure 3F). Consistently, the level of miR-1207-5p was also reduced in OS cell lines in comparison with that of the normal control Hob cells (Figure 3G). The correlation between the expression of miR-1207-5p and OR3A4 in OS tissues was analyzed with the Spearman test. As presented in Figure 3H, the expression of OR3A4 and miR-1207-5p was significantly negatively correlated in OS tissues. These data confirmed the negative role of OR3A4 in regulating the expression of miR-1207-5p in OS.
G6PD Was a Target of miR-1207-5p in OS
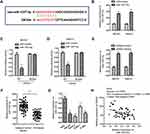
The function of miRNAs in cells mainly relies on modulating the expression of downstream targets. To further understand the role of miR-1207-5p in OS, the miR-1207-5p-targeted mRNAs were predicted with the miRDB database. Notably, the glucose-6-phosphate dehydrogenase (G6PD) was predicted as a target of miR-1207-5p. The possible complementary binding sites of miR-1207-5p at the 3ʹ-UTR of G6PD are shown in Figure 4A. Luciferase reporter assay showed that overexpression of miR-1207-5p significantly inhibited the luciferase activity of OS cells expressing WT 3ʹ-UTR of G6PD. However, no obvious inhibitory effect was found for the cells transfected with miR-1207-5p and mutated 3ʹ-UTR of G6PD (Figure 4B and C). To validate whether the binding of miR-1207-5p with the 3ʹ-UTR of G6PD affected the expression of G6PD, RT-qPCR analysis was performed to evaluate the mRNA level of G6PD in OS cells harboring miR-1207-5p mimics or miR-Control. The data uncovered that overexpressed miR-1207-5p markedly suppressed the mRNA level of G6PD (Figure 4D). Consistent with these findings, highly expressed miR-1207-5p decreased the protein level of G6PD in MG-63 and SaoS-2 cells (Figure 4E). These results indicated G6PD as a target of miR-1207-5p in OS.
As OR3A4 was a molecular sponge of miR-1207-5p, the effect of OR3A4 on the expression of G6PD was investigated. Both MG-63 and SaoS-2 cells were transfected with shRNA-OR3A4 or shRNA-control. The level of G6PD was detected by RT-qPCR and Western blot, respectively. The data showed that the down-regulation of OR3A4 markedly suppressed both the mRNA and protein levels of G6PD in OS cells (Figure 4F and G). These findings suggested that OR3A4 sponged miR-1207-5p and consequently regulated the level of G6PD in OS.
Down-Regulation of OR3A4 Inhibited the NADPH Production of OS Cells
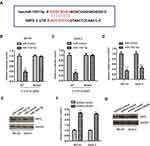
G6PD catalyzes the rate-limiting step of PPP, which generates NADPH and ribose 5-phosphate for nucleotide biosynthesis and glycolysis. Since OR3A4 modulated the expression of G6PD, the effect of OR3A4 on the production of NADPH was explored. As indicated in Figure 5A, the down-regulation of OR3A4 significantly reduced the level of NADPH in both MG-63 and SaoS-2 cells. Consistently, depletion of OR3A4 inhibited the glucose consumption and lactate production of OS cells (Figure 5B and C). These results suggested that knockdown of OR3A4 suppressed the glycolysis of OS cells. To further confirm whether G6PD mediated the function of OR3A4 in OS cells, both MG-63 and SaoS-2 cells were transfected with HA-tagged G6PD. The transfection of G6PD was validated by Western blot (Figure 5D). It is well documented that NADPH maintains the pool of reduced glutathione to balance the redox state (ROS).34 Since the production of NADPH was reduced with OR3A4 depletion, the level of ROS was measured with the down-regulation of OR3A4. Consistent with the reduced level of NADPH with shRNA-OR3A4, the intracellular production of ROS was significantly increased in OS cells (Figure 5E). Moreover, restoration of G6PD attenuated the up-regulated ROS level in OS cells (Figure 5E). Because the level of ROS was tightly associated with the endoplasmic reticulum (ER) stress, to investigate whether the increased level of ROS would induce ER stress of OS cells, the expression of PERK and phosphorylated IRE1, both ER stress markers, was detected. The results showed that the down-regulation of OR3A4 increased the abundance of these two markers, suggesting ER stress with depletion of OR3A4 (Figure 5F). Moreover, overexpression of G6PD attenuated the up-regulation of PERK and the activation of IRE1 (Figure 5F). Taken together, our results provided the possibility that the depletion of OR3A4 reduced the level of NADPH, which led to the accumulation of ROS and exacerbated ER stress-driven cell apoptosis. The proliferation of OS cells co-transfected with shRNA-OR3A4 and HA-G6PD was detected by CCK-8 assay. The data showed that overexpression of G6PD significantly reversed the growth defects of OS cells induced by OR3A4 depletion (Figure 5G and H). Collectively, these findings suggested that OR3A4 regulated the miR-1207-5p/G6PD axis and modulated the malignant behaviors of OS cells.
Discussion
Non-coding RNAs, including miRNAs, lncRNAs and circular RNAs, play important roles in the development of human cancers via regulating gene expression.6,7 Elucidating the underlying mechanism of lncRNAs in cancers might provide diagnostic and therapeutic strategies for the treatment of OS.7 In this study, we found that lncRNA OR3A4 was up-regulated in OS tissues and significantly correlated with the advanced progression of OS patients, which indicated the potential function of OR3A4 in OS.
LncRNA OR3A4 is generally considered as an oncogene and highly expressed in several types of cancers including ovarian cancer, hepatocellular carcinoma, breast cancer and gastric cancer.26,27,29 Overexpressed OR3A4 promoted the metastasis, angiogenesis, and drug resistance of cancer cells. Consistently, in the current study, OR3A4 was found overexpressed in OS tissues and cell lines. Knockdown of OR3A4 significantly inhibited the proliferation, colony formation and promoted the apoptosis of OS cells, suggesting the critical involvement of OR3A4 in modulating the malignant behaviors of OS cells. The ceRNA theory indicated that lncRNAs acted as a sponge of miRNAs to manipulate the expression of coding genes.35,36 Therefore, to further understand the mechanism of OR3A4 in OS, the binding miRNAs of OR3A4 were predicted. Interestingly, OR3A4 was found to bind miR-1207-5p and depletion of OR3A4 increased the expression of miR-1207-5p in OS cells. Notably, miR-1207-5p was down-regulated in OS tissues and negatively correlated with the level of OR3A4. These results were consistent with previous studies that miR-1207-5p was decreased and acted as a tumor suppressor in cancers, such as hepatocellular carcinoma, lung cancer and triple-negative breast cancer.37–39 Our findings demonstrated that OR3A4 regulated the OS progression via sponging miR-1207-5p.
A growing body of evidence has documented that dysregulation of miRNAs is closely associated with tumorigenesis via modulating the expression of target mRNAs. As OR3A4 decreased the level of miR-1207-5p in OS cells, the targets of miR-1207-5p were predicted to further characterize the mechanism of OR3A4 in OS. G6PD, the rate-limiting enzyme in PPP, was identified as the target of miR-1207-5p. Interestingly, previous studies showed that G6PD was overexpressed in a variety of cancers and predicted poor prognosis of cancer patients.32,33 G6PD is essential for the NADPH production and glycolysis of cancer cells. In this study, miR-1207-5p bound to the 3ʹ-UTR of G6PD and inhibited the expression of G6PD in OS cells. Consistent with the increased level of miR-1207-5p, the down-regulation of OR3A4 significantly reduced the level of G6PD in OS cells. Meanwhile, overexpression of G6PD reversed the decreased cell proliferation induced by OR3A4 depletion. Our results demonstrated that G6PD was targeted by miR-1207-5p and involved in the function of OR3A4 in the progression of OS.
In conclusion, our results illustrated that OR3A4 was overexpressed in OS tissues and regulated the growth of OS cells. Mechanistically, OR3A4 functioned as a molecular sponge to down-regulate miR-1207-5p, thereby resulting in the enhanced expression of G6PD. The findings suggested the potential suppressive role of OR3A4 in regulating the proliferation of OS cells via the OR3A4/miR-1207-5p/G6PD axis. Additionally, there are several questions remain to be answered regarding the function of OR3A4 in OS. Other the roles and mechanisms of OR3A4 in crucial signaling pathways during the progression of osteosarcoma need to be explored to enrich the functional mechanism of OR3A4 in OS. Our findings in this study provided preliminary insight into the suppressive role of OR3A4 in the proliferation of OS cells. More experiments, especially in vivo assays, are needed to investigate the function of OR3A4 in the pathogenesis of OS, which would increase the possibility that targeting OR3A4 as a potential strategy for the treatment of OS.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Xie L, Ji T, Guo W. Anti-angiogenesis target therapy for advanced osteosarcoma (Review). Oncol Rep. 2017;38(2):625–636. doi:10.3892/or.2017.5735
2. Wu G, Liang Q, Liu Y. Primary osteosarcoma of frontal bone: a case report and review of literature. Medicine (Baltimore). 2017;96(51):e9392. doi:10.1097/MD.0000000000009392
3. Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8(10):705–718.
4. Tian T, Wang M, Lin S, et al. The impact of lncRNA dysregulation on clinicopathology and survival of breast cancer: a systematic review and meta-analysis. Mol Ther Nucleic Acids. 2018;12:359–369. doi:10.1016/j.omtn.2018.05.018
5. Quan J, Pan X, Zhao L, et al. LncRNA as a diagnostic and prognostic biomarker in bladder cancer: a systematic review and meta-analysis. Onco Targets Ther. 2018;11:6415–6424. doi:10.2147/OTT
6. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
7. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
8. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi:10.1016/j.cell.2013.02.012
9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
10. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi:10.1055/s-00000069
11. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi:10.1146/annurev-biochem-060308-103103
12. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
13. Nugent M. microRNA and bone cancer. Adv Exp Med Biol. 2015;889:201–230.
14. Nugent M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer Manag Res. 2014;6:15–25. doi:10.2147/CMAR
15. Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: review. Front Pediatr. 2015;3:69. doi:10.3389/fped.2015.00069
16. Zhang J, Yan YG, Wang C, Zhang SJ, Yu XH, Wang WJ. MicroRNAs in osteosarcoma. Clin Chim Acta. 2015;444:9–17. doi:10.1016/j.cca.2015.01.025
17. Chang L, Shrestha S, LaChaud G, Scott MA, James AW. Review of microRNA in osteosarcoma and chondrosarcoma. Med Oncol. 2015;32(6):613. doi:10.1007/s12032-015-0613-z
18. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
19. Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21(41):11680–11687. doi:10.3748/wjg.v21.i41.11680
20. Yang C, Wu D, Gao L, et al. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7(12):13479–13490. doi:10.18632/oncotarget.7266
21. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi:10.3390/ijms19051310
22. Yu X, Hu L, Li S, et al. Long non-coding RNA Taurine upregulated gene 1 promotes osteosarcoma cell metastasis by mediating HIF-1alpha via miR-143-5p. Cell Death Dis. 2019;10(4):280. doi:10.1038/s41419-019-1509-1
23. Fei D, Sui G, Lu Y, Tan L, Dongxu Z, Zhang K. The long non-coding RNA-ROR promotes osteosarcoma progression by targeting miR-206. J Cell Mol Med. 2019;23(3):1865–1872. doi:10.1111/jcmm.2019.23.issue-3
24. Li W, Fu Q, Man W, Guo H, Yang P. LncRNA OR3A4 participates in the angiogenesis of hepatocellular carcinoma through modulating AGGF1/akt/mTOR pathway. Eur J Pharmacol. 2019;849:106–114. doi:10.1016/j.ejphar.2019.01.049
25. Zhong M, Wang WL, Yu DJ. Long non-coding RNA OR3A4 is associated with poor prognosis of human non-small cell lung cancer and regulates cell proliferation via up-regulating SOX4. Eur Rev Med Pharmacol Sci. 2019;23(15):6524–6530. doi:10.26355/eurrev_201908_18537
26. Guo FF, Jiang MM, Hong LL, et al. Long non-coding RNA OR3A4 promotes metastasis of ovarian cancer via inhibiting KLF6. Eur Rev Med Pharmacol Sci. 2019;23(6):2360–2365. doi:10.26355/eurrev_201903_17380
27. Liu G, Hu X, Zhou G. Long non-coding RNA OR3A4 promotes proliferation and migration in breast cancer. Biomed Pharmacother. 2017;96:426–433. doi:10.1016/j.biopha.2017.10.011
28. Shang J, Xu YD, Zhang YY, Li M. Long noncoding RNA OR3A4 promotes cisplatin resistance of non-small cell lung cancer by upregulating CDK1. Eur Rev Med Pharmacol Sci. 2019;23(10):4220–4225. doi:10.26355/eurrev_201905_17926
29. Guo X, Yang Z, Zhi Q, et al. Long noncoding RNA OR3A4 promotes metastasis and tumorigenicity in gastric cancer. Oncotarget. 2016;7(21):30276–30294. doi:10.18632/oncotarget.7217
30. Tiwari M. Glucose 6 phosphatase dehydrogenase (G6PD) and neurodegenerative disorders: mapping diagnostic and therapeutic opportunities. Gene Dis. 2017;4(4):196–203. doi:10.1016/j.gendis.2017.09.001
31. Cocco P. Does G6PD deficiency protect against cancer? A critical review. J Epidemiol Community Health. 1987;41(2):89–93. doi:10.1136/jech.41.2.89
32. Forteleoni G, Argiolas L, Farris A, Ferraris AM, Gaetani GF, Meloni T. G6PD deficiency and breast cancer. Tumori. 1988;74(6):665–667. doi:10.1177/030089168807400608
33. Wang J, Yuan W, Chen Z, et al. Overexpression of G6PD is associated with poor clinical outcome in gastric cancer. Tumour Biol. 2012;33(1):95–101. doi:10.1007/s13277-011-0251-9
34. Liu CL, Hsu YC, Lee JJ, et al. Targeting the pentose phosphate pathway increases reactive oxygen species and induces apoptosis in thyroid cancer cells. Mol Cell Endocrinol. 2020;499:110595. doi:10.1016/j.mce.2019.110595
35. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi:10.1002/jcp.27941
36. Cui X, Wang J, Guo Z, et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17(1):123. doi:10.1186/s12943-018-0877-y
37. Zhao G, Dong L, Shi H, et al. MicroRNA-1207-5p inhibits hepatocellular carcinoma cell growth and invasion through the fatty acid synthase-mediated Akt/mTOR signalling pathway. Oncol Rep. 2016;36(3):1709–1716. doi:10.3892/or.2016.4952
38. Dang W, Qin Z, Fan S, et al. miR-1207-5p suppresses lung cancer growth and metastasis by targeting CSF1. Oncotarget. 2016;7(22):32421–32432. doi:10.18632/oncotarget.v7i22
39. Hou X, Niu Z, Liu L, et al. miR-1207-5p regulates the sensitivity of triple-negative breast cancer cells to Taxol treatment via the suppression of LZTS1 expression. Oncol Lett. 2019;17(1):990–998. doi:10.3892/ol.2018.9687
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.