Back to Journals » Cancer Management and Research » Volume 12
lncRNA NR4A1AS Upregulates miR-221 Through Demethylation to Promote Cell Proliferation in Oral Squamous Cell Carcinoma
Authors Yang L, Li G, Gao Y, Ou N, Yu T, Ren S
Received 10 December 2019
Accepted for publication 12 June 2020
Published 2 July 2020 Volume 2020:12 Pages 5285—5292
DOI https://doi.org/10.2147/CMAR.S241769
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Liuqing Yang,1,2 Guanghui Li,3 Ya Gao,4 Nini Ou,1 Tingting Yu,5 Shirong Ren1
1Department of Stomatology, Ningbo Dental Hospital, Ningbo City, Zhejiang Province 315000, People’s Republic of China; 2Ningbo Institute of Oral Health, Ningbo City, Zhejiang Province 315000, People’s Republic of China; 3Department of Stomatology, Fujian Medical University, Fuzhou City, Fujian Province 350122, People’s Republic of China; 4Department of Stomatology, Shanghai First People’s Hospital, Shanghai 201600, People’s Republic of China; 5Department of Stomatology, Yinzhou Dental Hospital, Ningbo City, Zhejiang Province 315000, People’s Republic of China
Correspondence: Liuqing Yang
Department of Stomatology, Ningbo Dental Hospital, No. 1821 Ningchuan Road, Ningbo City, Zhejiang Province 315000, People’s Republic of China
Email [email protected]
Background: Cell proliferation of oral squamous cell carcinoma (OSCC) is precisely regulated with a cascade of genes and pathways. Previous studies have identified NR4A1 as an oncogene and plays a crucial role in colorectal cancer development and progression. This study was performed to investigate the potential interaction between lncRNA NR4A1AS and miR-221 and how their interaction is modulated in periodontitis.
Patients and Methods: Research subjects of this study included 62 OSCC patients. Cell transfection and RT-qPCR were applied to detect the expression levels of NR4A1AS and miR-221. Methylation-specific PCR (MSP) was carried out to determine the demethylation of miR-221 by NR4A1AS. CCK-8 assay was used to detect the proliferation of OSCC cells with the overexpression of NR4A1AS or/and overexpression of miR-221.
Results: In this study, we observed that NR4A1AS was upregulated in tumor tissue samples of OSCC, and its high expression levels were significantly correlated with poor survival in patients with OSCC. In addition, miR-221 was significantly down-regulated in OSCC tumors. NR4A1AS and miR-221 were significantly and positively correlated in OSCC tumors but not in non-dysplastic tissue. In OSCC cells, overexpression of NR4A1AS led to upregulation of miR-221 and decreased the methylation of miR-221 gene. However, overexpression of miR-221 did not affect the expression of NR4A1AS in OSCC cells. In addition, overexpression of NR4A1AS or miR-221 increased the proliferation rate of OSCC cells.
Conclusion: This study is the first to report that NR4A1AS is upregulated in OSCC. Moreover, we also propose that miR-221 is modulated by NR4A1AS through demethylation and the upregulation of NR4A1AS or miR-221 promotes the proliferation of OSCC cells, which suggests that anti-NR4A1AS might be a perspective approach for the therapy of OSCC.
Keywords: oral squamous cell carcinoma, lncRNA NR4A1AS, miR-221, proliferation
Introduction
Oral cancer is a common malignancy in clinical practices.1 According to the latest GLOBOCAN statistics, oral cancer affected 354,864 new cases, accounting for 2.0% of all cancer cases. In addition, 1.9% of all cancer deaths in 2018 were caused by oral cancer.2 Oral squamous cell carcinoma (OSCC) is a specific kind of oral cancer, which mainly invades the floor of mouth and tongue.3 Smoking, drinking as well as areca (betel) chewing are the major risk factors for OSCC.4–6 However, early diagnosis of OSCC is extremely difficult and considered as an effective strategy to increase survival rate of patients. Even though anti-cancer therapy has been developed over decades, OSCC patients without timely early-stage diagnosis still have low survival rates.7 Therefore, a precise and practicable diagnosis approach for early-stage OSCC is urgently needed.
The progression of OSCC is accompanied by the abnormally altered signaling transduction pathways of multiple molecules.8 Therefore, it is pivotal to understand the functions of different molecular pathways, which can provide a perspective insight into the development of OSCC therapies.9 It has been demonstrated that the molecular pathogenesis of OSCC also requires the participation of non-coding RNAs (ncRNAs), such as miRNA and long (>200 nt) ncRNAs (lncRNAs).10 lncRNAs were first reported to be involved in OSCC in 2013. Studies investigating the differential expression of lncRNAs in clinical cohorts of OSCC or tongue squamous cell carcinoma (TSCC) patients have identified several differentially expressed lncRNAs associated with the presence of disease or with clinic-pathological features including survival.11,12 Furthermore, the regulation of key ncRNA players might assist OSCC cells to promote or suppress cancer progression.13,14 However, the functions of most lncRNAs in OSCC are still unclear. Recently, lncRNA NR4A1AS was demonstrated to be an oncogenic lncRNA in colorectal cancer,15 while its role in OSCC is unknown. Our preliminary RNA-sequence analysis revealed that the expression of NR4A1AS in OSCC was abnormally changed, and NR4A1AS displayed an inverse correlation with miR-221, an oncogenic miRNA discovered in OSCC.16 This study was carried out to explore the potential interaction between miR-221 and NR4A1AS in OSCC and attempt to reveal the underlying mechanisms.
Patients and Methods
OSCC Patients and Specimens
Research subjects of this study were 62 OSCC patients (37 males and 25 females, 44 years to 68 years old, mean age 57.4 ± 6.2 years old) who were admitted to Ningbo Dental Hospital between April 2012 and April 2014. More information about clinical features of OSCC patients was described in Table 1. All patient specimen collection methods were reviewed and approved by the Ethics Review Committee of aforementioned hospital. Two surgeons participated in the fine needle biopsy operations for every patient recruited in our research. Patients’ inclusion criteria were: 1) newly diagnosed cases; 2) no therapies for any diseases were initiated within 3 months before this study; 3) no other systemic diseases were diagnosed. Exclusion criteria were: 1) recurrent cases; 2) multiple chronic diseases were diagnosed. All patients were informed of the experimental principle of this project and all of them signed the informed consent. All patients strictly followed follow-up strategy: 1) Recheck every three months within 2 years; 2) Recheck every half a year within 5 years; 3) After 5 years, recheck at any time when patients felt physically unpleasant. Paired OSCC and adjacent (within 3 cm around tumors) non-dysplastic tissues were collected from each patient through fine needle biopsies and their distinction with dysplastic tissue was distinguished by histopathological microscope. All tissue samples were stored in liquid nitrogen before use.
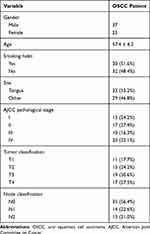 |
Table 1 Clinicopathological Data of Patients with OSCC |
Treatments and Follow-Up
Treatment strategies were approached mainly based on patients’ clinical stages and health conditions. According to AJCC staging criteria, 12 (stage I), 13 (stage II), 18 (stage III) and 19 (stage IV) OSCC patients were recruited from the day of admission. All 62 patients were followed up every month to record their survival, and all of them had completed follow-up strategies.
OSCC Cells and Cell Culture
Human OSCC cell lines SCC25 and SCC090 were purchased from ATCC (USA) and used in the following experiments. According to the instructions from ATCC, cells were cultivated in a medium composed of 90% Eagle’s Minimum Essential Medium and 10% fetal bovine serum (FBS) supplemented with 2 mM L-Glutamine. Cells were cultured at 37°C with 5% CO2. Cells were harvested at 5% confluence and used to perform subsequent transfections.
Transient Transfections
Human NR4A1AS sequence was amplified and subcloned into pcDNA3.1 vector at EcoRI and NotI sites to generate pcDNA3.1-NR4A1AS overexpression vector. Control mi-RNA and miR-221 mimic were obtained from Ji Kai Gene (Shanghai, China). SCC25 and SCC090 cells were transfected with 10 nM NR4A1AS overexpression vector or 40 nM miRNA using lipofectamine 2000 (Invitrogen) as transfection reagent. Control (C) cells were untransfected cells. Cells transfected with empty pcDNA3.1 or control miRNA were used as negative control (NC) cells. The sequence of miR-221 mimic was: 5′-AGCUACAUUGUCUGCUGGGUUUC-3′. The sequence of control miRNA was: 5′-UCAGCAUAGCAGCGUACCUAGCU-3′. All the following experiments were performed at 48 h post-transfection.
RNA Preparations
Trizol reagent (Invitrogen) was used to isolate total RNAs from tissue samples as well as SCC25 and SCC090 cells. To harvest miRNAs, 85% ethanol was used for washing and RNA precipitations. Genomic DNA was removed by DNase I digestion. NanoDrop™ 2000 spectrophotometers (Thermo Scientific) was used to measure RNA concentrations. RNA integrity was analyzed by 5% urine-PAGE gel.
Real-Time Quantitative PCR (RT-qPCR)
RNA samples with satisfactory quality were subjected to reverse transcription (RT) using Precision nanoScript2 Reverse Transcription Kit (PrimerDesign) to synthesize cDNAs. QuantiTect SYBR Green PCR Kit (Qiagen) was used to prepare qPCR reactions. The expression levels of NR4A1AS were measured with GAPDH as endogenous control. All-in-One™ miRNA qRT-PCR Reagent Kit (Genecopoeia) was applied to measure the expression levels of mature miR-221 with U6 as endogenous control. All PCR reactions were repeated 3 times and 2−ΔΔCT method was used to normalize gene expression. The primer sequences were as follows:
NR4A1AS forward: 5′-AGGGCTGCAAGGGCTTCT-3′,
NR4A1AS reverse: 5′-GGCAGATGTACTTGGCGTTTTT-3′,
GAPDH forward: 5′-CCATCCACAGTCTTCTGGGT-3′,
GAPDH reverse: 5′-GATCATCAGCAATGCCTCCT-3′,
MiR-221 forward: 5′-GGGAAGCTACATTGTCTGC-3′,
MiR-221 reverse: 5′-CAGTGCGTGTCGTGGAGT-3′,
U6 forward: 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3′,
U6 reverse: 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3′.
Methylation-Specific PCR (MSP)
SCC25 and SCC090 cells harvested at 48 h post-transfection were subjected to isolation of genomic DNA using Monarch® Genomic DNA Purification Kit (NEB). NanoDrop™ 2000 spectrophotometer was used to measure DNA concentrations. Genomic DNA was converted using EZ DNA Methylation-GoldTM kit (ZYMO RESEARCH). Taq 2X Master Mix (NEB) was used to prepare PCR reactions to analyze gene demethylation. The primers were designed by using Methylation Primer Express (Applied Biosystems). The sequences of primers used were:
Cell Counting Kit-8 (CCK-8) Assay
SCC25 and SCC090 cells harvested at 48 h post-transfection were subjected to cell proliferation assay by CCK-8 kit (Dojindo). At first, cells were counted and 3000 cells with 0.1 mL culture medium were planted to each well of 96-well plate. Cells were cultivated under aforementioned conditions and were collected every 24 h for a total of 4 d. CCK-8 solution was added to reach 10% final concentration at 4 h before cell collection. OD values were measured at 450 nm to reflect cell proliferation.
Statistical Analysis
Data of at least 3 biological replicates were expressed as mean ± SEM. Paired t test was used to compare differences between OSCC and non-dysplastic tissues of the same patient. ANOVA (one-way) and Tukey’s test were used to compare differences among multiple groups. Spearman correlation coefficient was used to analyze correlations. Sixty-two OSCC patients were sub-grouped into high or low NR4A1AS level group (n = 31) according to the RT-qPCR data. The cutoff value was defined by using the median value of the expression of NR4A1AS in OSCC tissues. Survival curves were plotted by Kaplan-Meier and compared by Log rank test. P < 0.05 was set to be statistically significant.
Results
Upregulation of NR4A1AS in OSCC Predicted Poor Survival
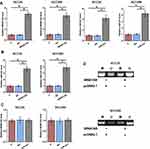
The expression of NR4A1AS in OSCC tumor or non-dysplastic samples from 62 OSCC patients were detected by performing RT-qPCR experiments. It was observed that the expression levels of NR4A1AS were significantly higher in OSCC tumor tissues compared to that in non-dysplastic tissues (Figure 1A, p < 0.05). Survival curves were plotted for both high and low NR4A1AS expression groups. The survival rate of patients with high expression levels of NR4A1AS was significantly lower than that of patients with low expression levels of NR4A1AS (Figure 1B), which suggested that high expression levels of NR4A1AS might be closely associated with poor survival in OSCC and possibly a negative factor in the pathogenesis of OSCC.
MiR-221 Was Also Upregulated in OSCC and Positively Correlated with NR4A1AS
Expression of miR-221 in OSCC tumor and non-dysplastic tumor samples from 62 OSCC patients was also detected by RT-qPCR. It was observed that the expression levels of miR-221 were significantly higher in OSCC tumor tissues compared to that in non-dysplastic tissues (Figure 2A, p < 0.05). Correlation analysis showed that the expression levels of miR-211 and NR4A1AS were significantly and positively correlated across OSCC tissues (Figure 2B), but not across non-dysplastic tissues (Figure 2C). The correlation between NR4A1AS and miR-211 indicated a regulatory mechanism underlying the relationship between these two molecular factors.
Overexpression of NR4A1AS Led to Upregulation of miR-221 and Induced Demethylation of miR-221 Gene in OSCC Cells
SCC25 and SCC090 cells were transfected with NR4A1AS overexpression vector or miR-221 mimic. Overexpression of NR4A1AS or miR-221 was confirmed by RT-qPCR at 48 h post-transfection (Figure 3A, p < 0.05). It was observed that overexpression of NR4A1AS led to a significant elevation of miR-221 in OSCC cells compared to that in C and NC groups (Figure 3B, p < 0.05). However, overexpression of miR-221 did not affect the expression of NR4A1AS (Figure 3C). MSP was performed to analyze the effects of overexpression of NR4A1AS on the demethylation of miR-221 gene. Compared to the transfection with empty pcDNA 3.1 vector, cells transfected with NR4A1AS overexpression vector showed a significant demethylation of miR-221 (Figure 3D), which suggested that the positive regulation of miR-221 by NR4A1AS might be due to this demethylation effect.
Overexpression of NR4A1AS or/and miR-221 Led to Increased Cell Proliferation Rate of OSCC Cells
The effects of overexpression of NR4A1AS or/and miR-221 on the proliferation of SCC25 and SCC090 cells were assessed by CCK-8 assay. Compared to non-transfection control group and empty vector transfection negative control group, overexpression of NR4A1AS or/and miR-221 induced dramatic increases in OSCC cell proliferation rate. However, there was no difference between the two control groups (C and NC) (Figure 4, p < 0.05). These results indicated the involvement of NR4A1AS and miR-221 in the pathogenesis of OSCC, and inhibiting the promotive roles of NR4A1AS or miR-221 in OSCC cell proliferation might be an effective approach for the therapy of OSCC.
Discussion
This study mainly aimed to explore the roles of NR4A1AS and miR-221 in OSCC pathogenesis, especially at the aspect of cell proliferation. We found that the expressions of NR4A1AS and miR-221 were both elevated in OSCC, and miR-221 upregulation might be modulated through demethylation by the overexpression of NR4A1AS.
The functionality of NR4A1AS has only been reported in colorectal cancer.15 It was shown that NR4A1AS was significantly upregulated in colorectal cancer, and silencing of NR4A1AS led to inhibited cancer cell proliferation, invasion and migration as well as promoted cancer cell apoptosis, indicating the important roles of NR4A1AS as an oncogenic lncRNA in colorectal cancer.15 However, the role of NR4A1AS in other diseases is unknown. Our study is the first to report the upregulation of NR4A1AS and its inverse correlation with miR-221 in OSCC. In addition, overexpression of NR4A1AS resulted in an increased cell proliferation rate in OSCC, which suggests NR4A1AS might be an oncogenic lncRNA in OSCC.
A major cause of high mortality of OSCC is the lack of sensitive early diagnosis marker for this disease.17 An accurate early diagnosis may provide a reliable guidance for the selection of treatment strategy for OSCC. Besides, there is no practical evaluation factor for the prognosis of OSCC patients. This study firstly demonstrated that NR4A1AS is overexpressed in tumor tissues of OSCC patients, which revealed that NR4A1AS might be a negative actor in the pathogenesis of OSCC. From our preliminary RNA-sequence analysis, we can obtain that the expression of NR4A1AS was significantly increased in OSCC tumors compared to normal tissues (Supplementary Figure S1). Moreover, it was also found that high expression levels of NR4A1AS were closely correlated with the poor survival of OSCC patients, indicating the potential clinical application of NR4A1AS as a prognostic evaluation factor for OSCC. MiR-221 is a well-established oncogenic miRNA in different types of cancer, including OSCC.18,19 MiR-221 not only regulates cancer cell behaviors in OSCC but also participates in the resistance of cancer cells to chemotherapies.18,19 The present study confirmed our previous RNA-sequence analysis showing the upregulation of miR-221 in OSCC (Supplementary Figure S1), and also revealed that miR-221 had a positive correlation with NR4A1AS. Furthermore, the overexpression of NR4A1AS could strongly elevates the expression levels of miR-221 in OSCC cell lines, but there was no effect of overexpression of miR-221 on the expression of NR4A1AS, which suggests that miR-221 did not affect degradation of NR4A1AS.
It has been reported that lncRNAs can regulate the expression of genes through methylation or demethylation.20,21 In this study, our MSP experiments proved that NR4A1AS could induce demethylation of miR-221 so as to upregulate the expression of miR-221 in OSCC. However, the underlying mechanism is still elusive and needs to be further explored. In order to clarify how NR4A1AS and miR-221 affect OSCC cell behaviors, the expression levels of NR4A1AS or/and miR-221 were elevated by in vitro transfection in OSCC cell lines. The results of CCK-8 assay showed that overexpression of NR4A1AS and miR-221 could promote cell proliferation. However, how and why NR4A1AS or miR-221 affect OSCC cell behaviors are still unknown. For the limitations of this study, firstly, we have not supplied any experimental evidence for the methylation site of miR-221 regulated by NR4A1AS to verify our findings in MSP experiments. Secondly, the underlying mechanism of how NR4A1AS or miR-221 affect OSCC cell behaviors is still a question and needs to be further explored.
In conclusion, the expression levels of NR4A1AS are strongly elevated as a negative regulator in OSCC, and the upregulation of miR-21 induced by overexpression of NR4A1AS might be through miR-21 demethylation. NR4A1AS and miR-21 promote the proliferation of OSCC cells and they might be considered as novel gene targets for OSCC therapy.
Abbreviations
OSCC, oral squamous cell carcinoma; ncRNAs, non-coding RNAs; lncRNAs¸ long (>200nt) ncRNAs.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of Ningbo Dental hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients and healthy volunteers provided written informed consent prior to their inclusion within the study.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Markopoulos AK. Current aspects on oral squamous cell carcinoma. Open Dent J. 2012;6:126–130. doi:10.2174/1874210601206010126
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Feller L, Lemmer J. Oral squamous cell carcinoma: epidemiology, clinical presentation and treatment. J Cancer Ther. 2012;3(04):263–268. doi:10.4236/jct.2012.34037
4. Ernani V, Saba NF. Oral cavity cancer: risk factors, pathology, and management. Oncology. 2015;89(4):187–195. doi:10.1159/000398801
5. Morse DE, Psoter WJ, Cleveland D, et al. Smoking and drinking in relation to oral cancer and oral epithelial dysplasia. Cancer Causes Control. 2007;18(9):919–929. doi:10.1007/s10552-007-9026-4
6. Lu HH, Kao SY, Liu TY, et al. Areca nut extract induced oxidative stress and upregulated hypoxia inducing factor leading to autophagy in oral cancer cells. Autophagy. 2010;6(6):725–737. doi:10.4161/auto.6.6.12423
7. Gómez I, Warnakulasuriya S, Varela Centelles PI, et al. Is early diagnosis of oral cancer a feasible objective? Who is to blame for diagnostic delay? Oral Dis. 2010;16(4):333–342. doi:10.1111/j.1601-0825.2009.01642.x
8. Rivera C, Venegas B. Histological and molecular aspects of oral squamous cell carcinoma. Oncol Lett. 2014;8(1):7–11. doi:10.3892/ol.2014.2103
9. Aggarwal S, John S, Sapra L, et al. Targeted disruption of PI3K/Akt/mTOR signaling pathway, via PI3K inhibitors, promotes growth inhibitory effects in oral cancer cells. Cancer Chemother Pharmacol. 2019;83(3):451–461. doi:10.1007/s00280-018-3746-x
10. Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143. doi:10.1016/j.cellsig.2017.10.009
11. Zhang L, Meng X, Zhu XW, et al. Long non-coding RNAs in oral squamous cell carcinoma: biologic function, mechanisms and clinical implications. Mol Cancer. 2019;18(1):102. doi:10.1186/s12943-019-1021-3
12. Pentenero M, Bowers L, Jayasinghe R, et al. World Workshop on Oral Medicine VII: functional pathways involving differentially expressed lncRNAs in oral squamous cell carcinoma. Oral Dis. 2019;25(Suppl 1):79–87. doi:10.1111/odi.13051
13. Arunkumar G, Murugan AK, Prasanna Srinivasa Rao H, et al. Long non-coding RNA CCAT1 is overexpressed in oral squamous cell carcinomas and predicts poor prognosis. Biomed Rep. 2017;6(4):455–462. doi:10.3892/br.2017.876
14. Yang YT, Wang YF, Lai JY, et al. Long non-coding RNA UCA 1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/β‐catenin signaling pathway. Cancer Sci. 2016;107(11):1581–1589. doi:10.1111/cas.13058
15. Xie X, Lin J, Liu J, et al. A novel lncRNA NR4A1AS up-regulates orphan nuclear receptor NR4A1 expression by blocking UPF1-mediated mRNA destabilization in colorectal cancer. Clin Sci (Lond). 2019;133(13):1457–1473. doi:10.1042/CS20181061
16. Sarkar S, Dubaybo H, Ali S, et al. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27kip1, p57kip2, and PUMA. Am J Cancer Res. 2013;3(5):465–477.
17. Carreras-Torras C, Gay-Escoda C. Techniques for early diagnosis of oral squamous cell carcinoma: systematic review. Med Oral Patol Oral Cir Bucal. 2015;20(3):e305–e315. doi:10.4317/medoral.20347
18. Zhou L, Jiang F, Chen X, et al. Downregulation of miR-221/222 by a microRNA sponge promotes apoptosis in oral squamous cell carcinoma cells through upregulation of PTEN. Oncol Lett. 2016;12(6):4419–4426. doi:10.3892/ol.2016.5250
19. Chen D, Yan W, Liu Z, et al. Downregulation of miR-221 enhances the sensitivity of human oral squamous cell carcinoma cells to Adriamycin through upregulation of TIMP3 expression. Biomed Pharmacother. 2016;77:72–78. doi:10.1016/j.biopha.2015.12.002
20. Liu H, Fang L, Cheng Y, et al. LncRNA PVT1 regulates prostate cancer cell growth by inducing the methylation of miR-146a. Cancer Med. 2016;5(12):3512–3519. doi:10.1002/cam4.900
21. Zhou L, Ren M, Zeng T, et al. TET2-interacting long noncoding RNA promotes active DNA demethylation of the MMP-9 promoter in diabetic wound healing. Cell Death Dis. 2019;10(11):813. doi:10.1038/s41419-019-2047-6
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.