Back to Journals » Cancer Management and Research » Volume 13
RETRACTED ARTICLE: LncRNA NOP14-AS1 Promotes Tongue Squamous Cell Carcinoma Progression by Targeting MicroRNA-665/HMGB3 Axis
Authors Li J, Fan S, Liu S, Yang G, Jin Q, Xiao Z
Received 21 November 2020
Accepted for publication 4 February 2021
Published 26 March 2021 Volume 2021:13 Pages 2821—2834
DOI https://doi.org/10.2147/CMAR.S293322
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
This paper has been retracted.
Jiayi Li,1 Shuxia Fan,2 Shuang Liu,3 Guang Yang,3 Qingsong Jin,3 Zhen Xiao3
1Department of Stomatology, The Third Affiliated Hospital of Qiqihar Medical University, Qiqihar, Heilongjiang, 161000, People’s Republic of China; 2Department of Stomatology, Qiqihaer Eye & ENT Hospital, Qiqihar, Heilongjiang, 161000, People’s Republic of China; 3Department of Stomatology, The First Hospital of Qiqihar (The Affiliated Qiqihar Hospital of Southern Medical University), Qiqihar, Heilongjiang, 161000, People’s Republic of China
Correspondence: Zhen Xiao
Department of Stomatology, The First Hospital of Qiqihar (The Affiliated Qiqihar Hospital of Southern Medical University), 30 Gongyuan Road, Qiqihar, Heilongjiang 161000, People’s Republic of China
Email [email protected]
Purpose: The expression profile, clinical effects, and detailed roles of NOP14 antisense RNA 1 (NOP14-AS1) in tongue squamous cell carcinoma (TSCC) remain ambiguous and need to be further explored. Thus, this work was initiated to offer further solid evidence regarding the expression and roles of NOP14-AS1 in TSCC. Furthermore, additional efforts were exerted to reveal the molecular events by which NOP14-AS1 affects the malignant behaviours of TSCC.
Methods: NOP14-AS1 expression was detected in TSCC tissues and cell lines using quantitative reverse transcription-polymerase chain reaction. Cell Counting Kit-8 assay, flow cytometric analysis, Transwell migration and invasion assays, and xenograft tumor model analysis were performed to assess the malignant biological behaviors of TSCC cells after NOP14-AS1 depletion. Mechanistic studies were performed using bioinformatics analysis, luciferase reporter assay, RNA immunoprecipitation, and rescue experiments.
Results: NOP14-AS1 upregulation was identified in TSCC tissues and cell lines. Patients with TSCC exhibiting a high NOP14-AS1 expression faced shorter overall survival than those with a low NOP14-AS1 expression. Functionally, NOP14-AS1 depletion facilitated apoptosis and impeded cell proliferation, migration, and invasion in TSCC. In vivo, the growth of TSCC cells was hindered by NOP14-AS1 depletion. Mechanically, NOP14-AS1 functioned as a competing endogenous RNA by sponging microRNA-665 (miR-665), thereby overexpressing the target high mobility group box 3 (HMGB3) of miR-665. Lastly, rescue experiments confirmed that the introduction of HMGB3 overexpression plasmid or miR-665 inhibitor could abrogate the inhibition of aggressive phenotypes triggered by NOP14-AS1 knockdown.
Conclusion: NOP14-AS1 executed pro-oncogenic activities in TSCC cells by targeting the miR-665/HMGB3 axis. The NOP14-AS1/miR-665/HMGB pathway may be a valuable prognostic indicator and therapeutic target for preventing TSCC.
Keywords: NOP14-AS1, tongue squamous cell carcinoma, high mobility group box 3, cancer therapy
Expression of Concern for this paper has been published.
Introduction
Cancer of the oral cavity and oropharynx is the eighth common malignant tumor occurring in humans worldwide.1 Tongue squamous cell carcinoma (TSCC) is the most common type of oral cancer and accounts for 25%–40% of all oral cancer cases worldwide.2 It is well known for unlimited growth and high incidence of metastasis, which give rise to disorders of speech, chewing, and swallowing and poor prognosis.3,4 Currently, the primary therapeutic strategies for TSCC that are widely accepted globally include surgical resection, radiotherapy, chemotherapy, and targeted therapy.5 In the past decades, there has been impressive progress in diagnosis and therapy technologies, thereby significantly improving the clinical efficiency of TSCC.6 However, long-term survival is unsatisfactory and unpredictable Insufficiency of detection methods and effective therapeutic techniques, distant metastasis, and recurrence are held accountable for the unfavorable prognosis.7 Consequently, complete recognition of TSCC pathogenesis may offer novel avenues for cancer diagnosis and therapy.
Long noncoding RNA (lncRNA) is a newly identified type of RNA transcripts comprising >200 nucleotides.8 They have limited protein-coding potential but have been considered a very hot research topic in biology.9–11 lncRNAs are engaged in a wide array of diverse physiological and pathological processes through gene regulation at the transcriptional, post-transcriptional, or translational level.12,13 Currently, lncRNAs have received considerable attention as novel critical controllers of cancer oncogenesis and progression.14 Aberrant lncRNA expression has been widely reported in TSCC, and their dysregulation is implicated in the regulation of numerous cellular processes of carcinogenesis.
MicroRNAs (miRNAs) represent one subgroup of short noncoding RNA transcripts managing gene expression through the induction of RNA-induced silencing complex (RISC) by base pairing with 3′-untranslated regions (3′-UTRs) of target genes.15 The dysregulation in miRNA has been widely reported in TSCC, which can play oncogenic or antioncogenic roles during TSCC genesis and progression.16 Recently, the competing endogenous RNA (ceRNA) theory has been proposed; lncRNAs can exert molecular decoys and scaffolds of miRNAs, thereby overexpressing the target genes of miRNAs.17,18 Thus, much hope is placed on exploring the dysregulated lncRNAs and miRNAs for developing novel therapeutic targets.
In recent years, the regulatory actions of lncRNAs in tumor biology have become the hotspot of research. To date, the expression status, clinical effects, and detailed roles of NOP14 antisense RNA 1 (NOP14-AS1) in TSCC remain ambiguous and need to be further explored. Therefore, NOP14-AS1 expression in TSCC tissue samples and cell lines was evaluated. Next, functional experiments were employed to assess the biological behaviors of NOP14-AS1 in TSCC. Importantly, the regulatory mechanisms of NOP14-AS1 were thoroughly clarified.
Materials and Methods
Ethics Approval and Consent to Participate
The current study was performed after obtaining approval from the Ethics Committee of The Third Affiliated Hospital of Qiqihar Medical University (approval number: EC.TAHQMU-2015.0112). All patients provided written informed consent before their enrolment in this research. The Animal Research Committee of The Third Affiliated Hospital of Qiqihar Medical University approved the animal experiments (approval number: ARC.TAHQMU-2019.0416), which were performed in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals.
Clinical Tissues
TSCC tissues and paired adjacent normal tissues were acquired from 56 patients with TSCC at The Third Affiliated Hospital of Qiqihar Medical University. Before surgery, radiochemotherapy or other anticancer therapies had never been administered in these patients. All tissues were stored in liquid nitrogen until further use.
Cell Culture and Transfection
Normal gingival epithelial cells (ATCC® PCS-200-014™) were acquired from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in minimum essential medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific). Dulbecco’s modified Eagle’s medium (DMEM; Gibco; Thermo Fisher Scientific) with 10% FBS was employed in the culture of the TSCC cell line CAL-27 (ATCC). The other two TSCC cell lines SCC-9 and SCC-15 were maintained in a 1:1 mixture of DMEM and Ham’s F12 medium (Gibco; Thermo Fisher Scientific) containing 10% FBS and 400 ng/mL hydrocortisone. All cells were incubated at 37°C in a humidified incubator supplied with 5% CO2.
Small interfering RNAs (siRNAs) for NOP14-AS1 (si-NOP14-AS1) and negative control (NC) siRNA (si-NC) were designed and offered by Shanghai GenePharma Co., Ltd. (Shanghai, China). The overexpressed high mobility group box 3 (HMGB3) plasmid pcDNA3.1-HMGB3 was constructed by RiboBio (Guangzhou, China). miRNA-665 (miR-665) mimic, NC mimic, miR-665 inhibitor, and NC inhibitor were also obtained from Shanghai GenePharma. TSCC cells were seeded into six-well plates, and transfection was performed by employing Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific).
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Trizol reagent (Invitrogen; Thermo Fisher Scientific) was employed for total RNA isolation. For NOP14-AS1 and HMGB3 quantification, complementary DNA was compounded with the PrimeScript® RT Reagent Kit (TaKaRa, Dalian, China) and then used for quantitative PCR using an SYBR® Premix Ex Taq™ II (TaKaRa). NOP14-AS1 and HMGB3 expression was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). For the expression analysis of miR-665, total RNA was reverse transcribed into cDNA using the miScript Reverse Transcription Kit (Qiagen GmbH, Hilden, Germany). Subsequently, PCR amplification was performed using the miScript SYBR Green PCR kit (Qiagen). U6 small nuclear RNA functioned as normalization for miR-665. The relative gene expression was analyzed based on the 2−ΔΔCq method.
Cell Cytoplasmic/Nuclear Fractionation
The separation of cytoplasmic and nuclear fractions in TSCC cells was performed using the Nuclear/Cytosol Fractionation Kit (Cell Biolabs, San Diego, CA, USA). After RNA extraction, qRT-PCR was performed to detect the relative distribution of NOP14-AS1 in the cytoplasmic and nuclear fractions. GAPDH and U6 small nuclear RNA served as the cytoplasmic and nuclear controls, respectively.
Cell Counting Kit-8 (CCK-8) Assay
After 24 h, transfected cells were seeded into a 96-well plate at a density of 1 × 103 per well. Cells were then cultivated for 0, 24, 48, and 72 h at 37°C with 5% CO2. To test cell proliferation, each well of the 96-well plate was loaded with 10 µL CCK-8 solution (Beyotime, Shanghai, China) and incubated at 37°C for 2 h. A microplate reader (Tecan Group Ltd., Mannedorf, Switzerland) was used to monitor the absorbance at 450 nm.
Flow Cytometric Analysis
The apoptosis of TSCC cells was detected using the Annexin V-FITC Apoptosis Detection Kit (KeyGen Biotech Co., Ltd., Nanjing, China). Approximately 1 × 106 cells were rinsed with phosphate-buffered saline (PBS) twice, and the supernatant fluid was removed. The collected cells were resuspended in 500 μL binding buffer. Later, 5 μL Annexin V-APC and 5 μL propidium iodide were introduced into the cell suspension, and additional culture was performed at room temperature for 15 min without light. Lastly, the late and early apoptotic rates were determined using flow cytometry (Becton Dickinson, San Jose, CA, USA).
Transwell Migration and Invasion Assays
Cell migration was assayed using the Transwell method. Briefly, TSCC cells were detached using trypsin and rinsed twice with PBS. The upper chambers of 24-well Transwell inserts were covered with approximately 5 × 104 cells resuspended in 200 μL FBS-free culture medium. A volume of 600 μL culture medium was added to the inferior cavity. Cells were allowed to pass through the porous membrane for 24 h. After the removal of cells remaining on the upper surface, the dyeing of migrated cells was performed using 0.1% crystal violet for 20 min at room temperature. Images of stained cells were captured using an inverted microscope (Olympus, Tokyo, Japan). Five images were chosen at random, and migrated cells that crossed the pores were counted. For the cell invasion assay, the same experimental steps were employed, except that the inserts were precoated with Matrigel (BD Biosciences).
Xenograft Tumor Model Analysis
The short hairpin RNA (shRNA) targeting NOP14-AS1 (sh-NOP14-AS1) and the NC shRNA (sh-NC) were designed and synthesized by Shanghai GenePharma Co., Ltd. The products were inserted into a pLVX-Puro lentiviral vector (Clontech, Palo Alto, CA, USA). The yield lentiviral vectors alongside psPAX2 and pMD2.G were introduced into 293T cells (Chinese Academy of Sciences, Shanghai, China) using ViaFect™ transfection reagent (Promega, Madison, WI, USA). After 2 days, the supernatant containing virus particles was harvested and used to infect CAL-27 cells. CAL-27 cells stably overexpressing sh-NOP14-AS1 or sh-NC were selected using puromycin.
Male BALB/c nude mice, aged 4 weeks, were acquired from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and subcutaneously injected with stably transfected CAL-27 cells. The width and length of tumor xenografts were recorded weekly. Tumor volume was calculated using the following formula: volume = 0.5 × length × width.2 Four weeks after inoculation, tumor xenografts were resected, imaged, and weighed.
Bioinformatics Analysis and Luciferase Reporter Assay
The potential miRNAs targeted NOP14-AS1 were predicted by ENCORI Starbase 3.0 (http://starbase.sysu.edu.cn/). TargetScan (http://www.targetscan.org/vert_60/) and miRDB tool (http://mirdb.org/miRDB/index.html) were used for searching miR-665’s target.
The wild-type (WT) and mutant (MUT) NOP14-AS1 fragments were amplified and inserted into the pmirGlO luciferase reporter vector (Promega), creating the WT-NOP14-AS1 and MUT-NOP14-AS1 vectors. The WT-HMGB3 and MUT-HMGB3 vectors were obtained employing the same experimental procedures. Regarding the reporter assay, the cotransfection of the constructed reporter vectors and miR-665 mimic or NC mimic in TSCC cells was achieved using Lipofectamine 2000. At 48 h after transfection, a dual-luciferase reporter assay system (Promega) was adopted for binding activity detection. Renilla luciferase activity was normalized to Firefly luciferase activity.
RNA Immunoprecipitation Assay
RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore, Bedford, MA, USA). Briefly, TSCC cells were lysed with RIPA lysis buffer. After centrifugation at 14,000 rpm and 4°C, 100 μL of the cell lysate was coprecipitated by probing with magnetic beads already conjugated with anti-Argonaute 2 (Ago2) antibody or normal IgG control (Millipore). After overnight incubation at 4°C, the coprecipitated magnetic beads were harvested and subjected to protein digestion via proteinase K. Ultimately, immunoprecipitated RNA was extracted and used in the subsequent qRT-PCR determination of NOP14-AS1 and miR-665. IgG served as the negative control, and input was regarded as the positive control.
Western Blot Analysis
Proteins were extracted using the RIPA buffer and quantified using the Enhanced BCA Protein Assay Kit (Beyotime). On 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, equal amounts of proteins were separated followed by transferring to polyvinylidene fluoride membranes. After soaking in 5% skim milk at room temperature for 2 h, primary antibodies specifically targeting HMGB3 (cat. no. ab32572; Abcam, Cambridge, UK) or GAPDH (cat. no. ab9485; Abcam) were added, and the membranes were incubated overnight at 4°C. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody (cat. no. ab6721; Abcam). Signals of the protein bands were visualized using Pierce™ ECL Western Blotting Substrate (Pierce; Thermo Fisher Scientific). GAPDH was used as the loading control.
Statistical Analysis
All experiments were conducted in three biological replicates, and the obtained data were shown as the mean ± standard deviation. Student’s t-test was applied for comparison of data between groups. One-way analysis of variance alongside Tukey’s post hoc test was used to compare the differences among multiple groups. Gene correlation analysis was performed via Pearson’s correlation coefficient. The Kaplan–Meier method was used to calculate overall survival and generate survival curves. The comparison between survival curves was performed using the Log rank test. SPSS 20.0 (IBM Corp., Armonk, NY, USA) was used for all statistical analyses. All patients with TSCC were classified into either low or high NOP14-AS1 groups based on the median value of NOP14-AS1 in TSCC tissues. P < 0.05 was considered to indicate statistical significance.
Results
NOP14-AS1 Downregulation Suppresses TSCC Cell Proliferation, Migration, and Invasion but Induces Cell Apoptosis in vitro
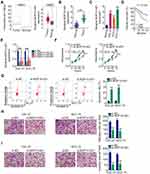
By searching The Cancer Genome Atlas (TCGA), NOP14-AS1 expression was examined in head and neck squamous cell carcinoma (HNSC) tissues and normal tissues. Figure 1A revealed that NOP14-AS1 expression increased evidently in HNSC tissues compared with normal tissues. NOP14-AS1 expression was then determined in 56 pairs of TSCC tissues and adjacent normal tissues. TSCC tissues possessed a higher NOP14-AS1 level than adjacent normal tissues (Figure 1B). Consistently, NOP14-AS1 was overexpressed in TSCC cell lines compared with that in normal gingival epithelial cells (Figure 1C). Furthermore, TSCC patients with high NOP14-AS1 expression faced shorter overall survival period than those with low NOP14-AS1 expression (Figure 1D).
Relatively higher NOP14-AS1 expression was detected in the CAL-27 and SCC-15 cell lines, which were thus selected as subjects of subsequent experiments. To ascertain whether NOP14-AS1 is involved in the tumorigenesis of TSCC, si-NOP14-AS1 or si-NC was transfected into CAL-27 and SCC-15 cells to knockdown endogenous NOP14-AS1 expression. NOP14-AS1 knockdown was verified by qRT-PCR (Figure 1E). si-NOP14-AS1#1 presented higher silencing efficiency; thus, siRNA was used in the following experiments. The effects of NOP14-AS1 on TSCC cell proliferation and apoptosis were assessed by CCK-8 assay and flow cytometric analysis, respectively. Transfection with si-NOP14-AS1 caused an obvious decrease in proliferation rate (Figure 1F) and an increase in apoptosis rate (Figure 1G) in CAL-27 and SCC-15 cells. Furthermore, the migratory (Figure 1H) and invasive (Figure 1I) properties of CAL-27 and SCC-15 cells were strikingly hindered by si-NOP14-AS1 treatment in contrast to si-NC treatment. Overall, the observations mentioned above supported the conclusion that NOP14-AS1 acts as an oncogenic lncRNA in TSCC.
miR-665 is Sponged by NOP14-AS1 in TSCC
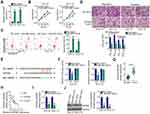
Next, the subcellular localization of NOP14-AS1 was explored to further elucidate the molecular events underlying the tumor-promoting roles of NOP14-AS1 in TSCC cells. Most NOP14-AS1 was noted to be enriched in the cytoplasm of TSCC cells (Figure 2A), which offered a theoretical basis for the ceRNA theory. Using ENCORI Starbase 3.0, a total of 72 miRNAs were predicted to contain putative binding sites within NOP14-AS1. Next, candidates from ENCORI Starbase 3.0 were intersected with the TCGA-HNSC database. As shown in Figure 2B, a total of 17 miRNAs were downregulated and selected for subsequent experimental confirmation. With the objective of corroborating miRNAs that could be affected by NOP14-AS1, their expression in NOP14-AS1-deficient TSCC cells was studied using qRT-PCR. miR-665 expression was enhanced in CAL-27 and SCC-15 cells transfected with si-NOP14-AS1, whereas the levels of other miRNAs remained unchanged (Figure 2C). Furthermore, downregulated miR-665 was found in TSCC tissues (Figure 2D), which exhibited an inverse expression relation with NOP14-AS1 (Figure 2E). To further confirm the direct relationship between NOP14-AS1 and miR-665 (Figure 2F), CAL-27 and SCC-15 cells were transfected with miR-665 mimic or NC mimic alongside WT-NOP14-AS1 or MUT-NOP14-AS1, and then luciferase reporter assay was performed. The results confirmed that the luciferase activity of WT-NOP14-AS1 instead of MUT-NOP14-AS1 was evidently decreased by miR-665 upregulation in CAL-27 and SCC-15 cells (Figure 2G). Additionally, the outcomes of RIP assay affirmed that both NOP14-AS1 and miR-665 could be remarkably enriched by anti-Ago2 in CAL-27 and SCC-15 cells compared with the anti-IgG NC group (Figure 2H). To sum up, NOP14-AS1 operates as a miR-665 sponge in TSCC cells.
miR-665 Directly Targets HMGB3 in TSCC Cells
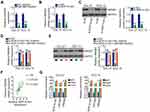
With results verifying miR-665 downregulation in TSCC, we next attempted to ascertain the roles of miR-665 in TSCC cells. miR-665 mimic was transfected into CAL-27 and SCC-15 cells, with NC mimic as the control. qRT-PCR analysis demonstrated that miR-665 expression remarkably increased after miR-665 mimic transfection (Figure 3A). Exogenous miR-665 expression notably attenuated proliferation (Figure 3B) but accelerated apoptosis (Figure 3C) in CAL-27 and SCC-15 cells. Through Transwell migration and invasion assays, a significant impairment of CAL-27 and SCC-15 cell migration and invasion (Figure 3D) was observed after miR-665 upregulation.
To determine the mechanism of action of miR-665 in TSCC, bioinformatics analysis was used to search for the target of miR-665. HMGB3 was predicted to harbor putative binding sequences for miR-665 (Figure 3E) and was selected for further identification considering its critical roles during tumor genesis and development.19–21 Next, luciferase reporter assay was performed. It was found that miR-665 mimic clearly lessened the luciferase activity of WT-HMGB3, whereas no evident suppression was identified in CAL-27 and SCC-15 cells when the binding sequences were mutated (Figure 3F). In addition, HMGB3 expression was high in TSCC tissues compared with that in adjacent normal tissues (Figure 3G). More intriguingly, Pearson’s correlation analysis illustrated an inverse expression correlation between miR-665 and HMGB3 in TSCC tissues (Figure 3H). Furthermore, upregulating miR-665 attenuated the HMGB3 mRNA (Figure 3I) and protein (Figure 3J) levels in CAL-27 and SCC-15 cells. Collectively, miR-665 directly targets HMGB3 in TSCC cells.
NOP14-AS1 Silencing Represses HMGB3 Expression in TSCC Cells by Sponging miR-665
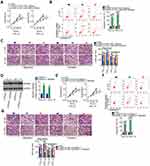
After validating HMGB3 as a target of miR-665, whether HMGB3 could be regulated by NOP14-AS1 in TSCC cells was explored. CAL-27 and SCC-15 cells were transfected with si-NOP14-AS1 and miR-665 inhibitor either alone or in combination. The silencing efficiency of miR-665 inhibitor in TSCC cells was affirmed by qRT-PCR. The delivery of miR-665 inhibitor caused a prominent downregulation of miR-665 in CAL-27 and SCC-15 cells (Figure 4A). NOP14-AS1 downregulation triggered an obvious inhibitory effect on the endogenous HMGB3 mRNA (Figure 4B) and protein (Figure 4C) levels in CAL-27 and SCC-15 cells, whereas the delivery of miR-665 inhibitor counteracted such effect (Figure 4D and E). Furthermore, HMGB3 expression was positively correlated with NOP14-AS1 expression in TSCC tissues (Figure 4F). Besides, NOP14-AS1, miR-665, and HMGB3 were preferentially concentrated in anti-Ago2 precipitates compared with that in the anti-IgG NC group (Figure 4G). On all accounts, these results confirmed that NOP14-AS1 acts as a ceRNA for miR-665 in TSCC cells, causing HMGB3 overexpression.
NOP14-AS1 Promotes the Oncogenicity of TSCC Cells by Targeting the miR-665/HMGB3 Axis
To validate that NOP14-AS1 aggravates the malignancy of TSCC by regulating the miR-665/HMGB3 axis, rescue experiments were performed by cotransfecting si-NOP14-AS1 and miR-665 inhibitor or NC inhibitor into TSCC cells. Thereafter, CCK-8 assay and flow cytometric analysis showed that the descending proliferation (Figure 5A) and ascending apoptosis (Figure 5B) imposed by NOP14-AS1 depletion were reversed by miR-665 inhibition. Additionally, the migratory and invasive (Figure 5C) capacities were strikingly hindered in NOP14-AS1-deficient CAL-27 and SCC-15 cells but largely recovered after miR-665 inhibitor cotransfection.
HMGB3 overexpression plasmid pcDNA3.1-HMGb3 was synthesized and transfected into TSCC cells to increase endogenous HMGB3 expression (Figure 5D). Similarly, the suppressed cell proliferation (Figure 5E) and promoted cell apoptosis (Figure 5F) triggered by si-NOP14-AS1 was offset by HMGB3 upregulation. Furthermore, restoring HMGB3 expression abolished the descending tendency of migration and invasion (Figure 5G) caused by NOP14-AS1 interference. Consequently, NOP14-AS1 interference decreases HMGB3 expression and attenuates the malignant properties of TSCC cells in vitro by sponging miR-665.
NOP14-AS1 Interference Inhibits TSCC Tumor Growth in vivo
To illustrate the influence of NOP14-AS1 on TSCC tumorigenesis, xenograft tumor model analysis was also conducted. Initially, NOP14-AS1 expression was determined in CAL-27 cells stably transfected with sh-NOP14-AS1 or sh-NC. qRT-PCR analysis indicated that NOP14-AS1 expression was significantly downregulated in CAL-27 cells infected with sh-NOP14-AS1 lentiviruses (Figure 6A), indicating that sh-NOP14-AS1 lentiviruses-infected CAL-27 cells could be used in the xenograft tumor model. Tumor xenografts infected with sh-NOP14-AS1 presented smaller tumor volume (Figure 6B and C) and lighter tumor weight (Figure 6D) than tumor xenografts in the sh-NC group. Next, the NOP14-AS1, miR-665, and HMGB3 expression levels in the tumor xenografts derived from CAL-27 cells transfected with sh-NOP14-AS1 or sh-NC were further tested. Decreased NOP14-AS1 (Figure 6E), increased miR-665 (Figure 6F), and lessened HMGB3 (Figure 6G) levels were identified in NOP14-AS1-depleted tumor xenografts. Altogether, in vivo data reconfirmed the pro-oncogenic actions of NOP14-AS1 in TSCC tumorigenesis.
Discussion
Several studies have uncovered that differentially expressed lncRNAs are important regulators in TSCC oncogenesis and progression.22–24 Accordingly, delving into the detailed roles of lncRNAs in TSCC is of pivotal importance for seeking novel targets of auxiliary diagnosis or therapy. Although >95,000 lncRNAs have been identified,25 only a handful of lncRNAs have been studied in detail in TSCC. Hence, this work was initiated to offer further solid evidence regarding the expression and roles of NOP14-AS1 in TSCC. Furthermore, the molecular events by which NOP14-AS1 affects the malignant behaviors of TSCC were elucidated in greater depth.
An increasing number of studies have focused on the expression status and specific roles of lncRNAs in TSCC. For instance, PART1,26 FALEC,27 and LINC0096128 were weakly expressed in TSCC and performed antioncogenic roles. In contrast, UCA1,29 PHACTR2-AS1,30 and FOXD2-AS131 were expressed at high levels in TSCC and performed pro-oncogenic actions during TSCC progression. To date, the expression, clinical value, and biological roles of NOP14-AS1 in TSCC progression remain elusive. In this work, NOP14-AS1 upregulation was observed in TSCC tissues and cell lines. High NOP14-AS1 expression was correlated with poor prognosis in patients with TSCC. Functionally, NOP14-AS1 interference facilitated apoptosis and impeded cell proliferation, migration, and invasion in TSCC. In vivo, the growth of TSCC cells was hindered by NOP14-AS1 depletion. Overall, these results may aid in the identification of promising targets for TSCC diagnosis and targeted treatment.
The mechanisms responsible for NOP14-AS1-mediated oncogenic activities in TSCC were further unveiled. lncRNAs in the nucleus participate in RNA processing, transcription, and chromatin interactions.32 Growing evidence illuminated that cytoplasmic lncRNAs could operate as ceRNAs for specific miRNAs, thereby upregulating the target mRNAs at the post-transcriptional level.33–35 Using cell cytoplasmic/nuclear fractionation assay, most NOP14-AS1 was identified to be located in the cytoplasm of TSCC cells, implying that lncRNAs may execute its roles via the ceRNA theory. By searching the ENCORI Starbase 3.0, NOP14-AS1 was found to contain theoretical binding sites of miR-665. Consistent with a previous study,36 miR-665 was found to be weakly expressed in TSCC and exerted cancer-suppressing roles. Further correlation analysis corroborated a negative correlation between the abundance of miR-665 and NOP14-AS1 in TSCC tissues. Additionally, NOP14-AS1 interference caused a notable miR-665 overexpression in TSCC cells. Importantly, using luciferase reporter and RIP assays, NOP14-AS1 and miR-665 were confirmed to exist in the same RISC.
Regarding the working mechanisms of miRNAs, they are capable of directly binding to the 3′-UTR of their target gene, thereby functioning as gene regulators.16 As a bioinformatics prediction, luciferase reporter assay and molecular analysis indicated that HMGB3 is a direct target of miR-665 for influencing the malignancy of TSCC cells. After identifying NOP14-AS1 as a miR-665 sponge, additional efforts were made to determine the interactions among NOP14-AS1, miR-665, and HMGB3 in TSCC. These data showed that NOP14-AS1 depletion lowered the HMGB3 mRNA and protein levels in TSCC cells, and miR-665 inhibition recovered the downregulated HMGB3 expression imposed by NOP14-AS1 silencing. Lastly, the coexistence of NOP14-AS1, miR-665, and HMGB3 in the same RISC was further certified by RIP assay. Consequently, NOP14-AS1 served as a ceRNA by sequestering miR-665 and then overexpressing HMGB3, forming a new ceRNA network in TSCC.
HMGB3 is a member of the high mobility group box subfamily, including HMGB1, HMGB2, HMGB3, and HMGB4.37 A couple of miRNAs have been revealed to be involved in controlling HMGB3 expression in human cancers;38–40 however, the relationship between miRNA and HMGB3 remains unaddressed in TSCC. Herein, it was verified that HMGB3 was negatively regulated by miR-665 in TSCC cells but positively modulated by NOP14-AS1. More intriguingly, HMGB3 manifested a negative expression correlation with miR-665 but a positive correlation with NOP14-AS1 in TSCC tissues. Ultimately, rescue experiments conducted on TSCC cells affirmed that the introduction of HMGB3 overexpression plasmid or miR-665 inhibitor could antagonize the suppression of aggressive phenotypes triggered by NOP14-AS1 knockdown. On all accounts, these observations, for the first time, presented evidence suggesting that NOP14-AS1 executed pro-oncogenic activities in TSCC cells by targeting the miR-665/HMGB3 axis.
Our study had several limitations. First, the sample size is small. Second, we only determined the effect of NOP14-AS1 depletion on the tumor growth of TSCC cells in vivo; yet, the impact of NOP14-AS1 silencing on TSCC tumor metastasis in vivo was not illustrated. Finally, loss-of-function assays were implemented on TSCC cells,whereas gain-of-function assays were not performed. We will resolve these limitations in the near future.
Conclusion
To sum up, NOP14-AS1 was extremely overexpressed in TSCC and closely related to worst overall survival in patients. NOP14-AS1 worked as a ceRNA for miR-665 and thus upregulated HMGB3, which ultimately promoted the oncogenicity of TSCC cells. Thus, the NOP14-AS1/miR-665/HMGB axis may exert important roles in TSCC progression, and further research is needed to explore whether the aix may be a valuable prognostic indicator and therapeutic target for preventing TSCC.
Funding
This study was supported by the Qiqihar Science & Technology Bureau (SFGG-201522).
Disclosure
The authors declare that they have no competing interests.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Irimie AI, Ciocan C, Gulei D, et al. Current insights into oral cancer epigenetics. Int J Mol Sci. 2018;19:3. doi:10.3390/ijms19030670
3. Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26(3–4):645–662. doi:10.1007/s10555-007-9082-y
4. Kimple AJ, Welch CM, Zevallos JP, Patel SN. Oral cavity squamous cell carcinoma–an overview. Oral Health Dent Manag. 2014;13(3):877–882.
5. Ren ZH, Wu HJ, Zhang S, et al. A new surgical strategy for treatment of tongue squamous cell carcinoma based on anatomic study with preliminary clinical evaluation. J Craniomaxillo Facial Surg. 2015;43(8):1577–1582. doi:10.1016/j.jcms.2015.07.034
6. Miller C, Shay A, Tajudeen B, et al. Clinical features and outcomes in young adults with oral tongue cancer. Am J Otolaryngol. 2019;40(1):93–96. doi:10.1016/j.amjoto.2018.09.022
7. Blatt S, Kruger M, Ziebart T, et al. Biomarkers in diagnosis and therapy of oral squamous cell carcinoma: a review of the literature. J Craniomaxillo Facial Surg. 2017;45(5):722–730. doi:10.1016/j.jcms.2017.01.033
8. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
9. Zhou C, Duan S. The role of long non-coding RNA NNT-AS1 in neoplastic disease. Cancers. 2020;12:11. doi:10.3390/cancers12113086
10. Galvao M, Coimbra EC. Long noncoding RNAs (lncRNAs) in cervical carcinogenesis: new molecular targets, current prospects. Crit Rev Oncol Hematol. 2020;156:103111. doi:10.1016/j.critrevonc.2020.103111
11. Saw PE, Xu X, Chen J, Song EW. Non-coding RNAs: the new central dogma of cancer biology. Sci China Life Sci. 2020.
12. Tu C, Yang K, Wan L, et al. The crosstalk between lncRNAs and the Hippo signalling pathway in cancer progression. Cell Prolif. 2020;53(9):e12887. doi:10.1111/cpr.12887
13. Javed Z, Khan K, Sadia H, et al. LncRNA & Wnt signaling in colorectal cancer. Cancer Cell Int. 2020;20(1):326. doi:10.1186/s12935-020-01412-7
14. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
15. Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. doi:10.2174/138920210793175895
16. Karatas OF, Oner M, Abay A, Diyapoglu A. MicroRNAs in human tongue squamous cell carcinoma: from pathogenesis to therapeutic implications. Oral Oncol. 2017;67:124–130. doi:10.1016/j.oraloncology.2017.02.015
17. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi:10.1002/jcp.27941
18. Shuwen H, Qing Z, Yan Z, Xi Y. Competitive endogenous RNA in colorectal cancer: a systematic review. Gene. 2018;645:157–162. doi:10.1016/j.gene.2017.12.036
19. Zhuang S, Yu X, Lu M, Li Y, Ding N, Ding Y. High mobility group box 3 promotes cervical cancer proliferation by regulating Wnt/beta-catenin pathway. J Gynecol Oncol. 2020;31(6):e91. doi:10.3802/jgo.2020.31.e91
20. Gu J, Xu T, Huang QH, Zhang CM, Chen HY. HMGB3 silence inhibits breast cancer cell proliferation and tumor growth by interacting with hypoxia-inducible factor 1alpha. Cancer Manag Res. 2019;11:5075–5089. doi:10.2147/CMAR.S204357
21. Zheng W, Yang J, Dong Z, et al. High mobility group box 3 as an emerging biomarker in diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:5979–5989. doi:10.2147/CMAR.S181742
22. Chen J, Liu L, Cai X, Yao Z, Huang J. Progress in the study of long noncoding RNA in tongue squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2020;129(1):51–58. doi:10.1016/j.oooo.2019.08.011
23. Wang J, Li L, Wu K, et al. Knockdown of long noncoding RNA urothelial cancer-associated 1 enhances cisplatin chemosensitivity in tongue squamous cell carcinoma cells. Die Pharmazie. 2016;71(10):598–602. doi:10.1691/ph.2016.6625
24. Yu J, Liu Y, Guo C, et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8(4):523–530. doi:10.7150/jca.17510
25. Xie C, Yuan J, Li H, et al. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42(Databaseissue):D98–103. doi:10.1093/nar/gkt1222
26. Zhao X, Hong Y, Cheng Q, Guo L. LncRNA PART1 exerts tumor-suppressive functions in tongue squamous cell carcinoma via miR-503-5p. Onco Targets Ther. 2020;13:9977–9989. doi:10.2147/OTT.S264410
27. Jia B, Xie T, Qiu X, et al. Long noncoding RNA FALEC inhibits proliferation and metastasis of tongue squamous cell carcinoma by epigenetically silencing ECM1 through EZH2. Aging. 2019;11(14):4990–5007. doi:10.18632/aging.102094
28. Zhang L, Shao L, Hu Y. Long noncoding RNA LINC00961 inhibited cell proliferation and invasion through regulating the Wnt/beta-catenin signaling pathway in tongue squamous cell carcinoma. J Cell Biochem. 2019;120(8):12429–12435. doi:10.1002/jcb.28509
29. Shi TT, Li R, Zhao L. Long noncoding RNA UCA1 regulates CCR7 expression to promote tongue squamous cell carcinoma progression by sponging miR-138-5p. Neoplasma. 2020;67(6):1256–1265. doi:10.4149/neo_2020_191119N1187
30. Yuan F, Miao Z, Chen W, et al. Long non-coding RNA PHACTR2-AS1 promotes tongue squamous cell carcinoma metastasis by regulating Snail. J Biochem. 2020;168(6):651–657. doi:10.1093/jb/mvaa082
31. Zhou G, Huang Z, Meng Y, Jin T, Liang Y, Zhang B. Upregulation of long non-coding RNA FOXD2-AS1 promotes progression and predicts poor prognosis in tongue squamous cell carcinoma. J Oral Pathol Med. 2020;49(10):1011–1018. doi:10.1111/jop.13074
32. Guo CJ, Xu G, Chen LL. Mechanisms of long noncoding RNA nuclear retention. Trends Biochem Sci. 2020;45(11):947–960. doi:10.1016/j.tibs.2020.07.001
33. Niu ZS, Wang WH, Dong XN, Tian LM. Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World j Gastroenterol. 2020;26(29):4240–4260. doi:10.3748/wjg.v26.i29.4240
34. Wu Y, Qian Z. Long non-coding RNAs (lncRNAs) and microRNAs regulatory pathways in the tumorigenesis and pathogenesis of glioma. Discov Med. 2019;28(153):129–138.
35. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20:22.
36. Wei T, Ye P, Yu GY, Zhang ZY. Circular RNA expression profiling identifies specific circular RNAs in tongue squamous cell carcinoma. Mol Med Rep. 2020;21(4):1727–1738. doi:10.3892/mmr.2020.10980
37. Nemeth MJ, Curtis DJ, Kirby MR, et al. Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. 2003;102(4):1298–1306. doi:10.1182/blood-2002-11-3541
38. Liu J, Wang L, Li X. HMGB3 promotes the proliferation and metastasis of glioblastoma and is negatively regulated by miR-200b-3p and miR-200c-3p. Cell Biochem Funct. 2018;36(7):357–365. doi:10.1002/cbf.3355
39. Li X, Wu Y, Liu A, Tang X. MiR-27b is epigenetically downregulated in tamoxifen resistant breast cancer cells due to promoter methylation and regulates tamoxifen sensitivity by targeting HMGB3. Biochem Biophys Res Commun. 2016;477(4):768–773. doi:10.1016/j.bbrc.2016.06.133
40. Chen X, Zhao G, Wang F, et al. Upregulation of miR-513b inhibits cell proliferation, migration, and promotes apoptosis by targeting high mobility group-box 3 protein in gastric cancer. Tumour Biol. 2014;35(11):11081–11089. doi:10.1007/s13277-014-2405-z
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.