Back to Journals » Cancer Management and Research » Volume 12
LncRNA NEAT1/miR-204/NUAK1 Axis is a Potential Therapeutic Target for Non-Small Cell Lung Cancer
Authors Zhao MM, Ge LY, Yang LF, Zheng HX, Chen G, Wu LZ, Shi SM, Wang N, Hang YP
Received 18 August 2020
Accepted for publication 23 November 2020
Published 29 December 2020 Volume 2020:12 Pages 13357—13368
DOI https://doi.org/10.2147/CMAR.S277524
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Ming-Ming Zhao, Lin-Yang Ge, Liang-Feng Yang, Hai-Xia Zheng, Gang Chen, Li-Zheng Wu, Shao-Ming Shi, Nan Wang, Yan-Ping Hang
Department of Respiratory and Critical Care Medicine, People’s Hospital of Gaochun, Nanjing 211300, People’s Republic of China
Correspondence: Yan-Ping Hang
Department of Respiratory and Critical Care Medicine, People’s Hospital of Gaochun, No. 53, Maoshan Road, Nanjing 211300, People’s Republic of China
Tel +86 25-57311232
Email [email protected]
Background: Long non-coding RNA (lncRNA) is a key part of non-coding RNA, and more and more evidence has revealed that it plays a vital role in tumors. NEAT1 is a lncRNA discovered in the early stage. However, it is still unclear whether NEAT1 and miR-204 play a regulatory role in lung cancer (LC). This research aimed to determine the biological function of NEAT1/miR-204 in non-small cell lung cancer (NSCLC).
Materials and Methods: In order to research the function of NEAT1 in NSCLC, RT-PCR, Western blot, luciferase assay and RNA immunoprecipitation assay were used to determine the relationship between NEAT1, miR-204 and NUAK1. CCK8 test, cell migration and invasion test were used to explore the influence of NEAT1 on proliferation and metastasis of LC cells. Tumor allotransplantation was used to detect the influence of NEAT1 on the growth of LC.
Results: The results revealed that NEAT1 was obviously enhanced in LC cell lines. Further functional analysis showed that low expression of NEAT1 obviously suppressed the growth, migration and invasion of NSCLC and facilitated cell apoptosis. Determination of luciferase reporter gene revealed that miR-204 was the direct target of NEAT1 in LC. In addition, NUAK1 was called the direct target of miR-204, and miR-204/NUAK1 had saved the role of NEAT1 in NSCLC cells. Tumor allotransplantation experiments showed that knocking down NEAT1 could inhibit the growth of LC.
Conclusion: In summary, our results showed that the down-regulation of NEAT1 in NSCLC inhibited its growth, migration and invasion through the miR-204/NUAK1 axis.
Keywords: NEAT1, miR-204, NUAK1, non-small cell lung cancer
Introduction
Lung cancer (LC), as the most common malignant respiratory tumor clinically, is still the main reason of tumor-related morbidity and mortality all over the world.1 According to the global statistics in 2018,2 there are more than 2,000,000 new patients with LC and 1,700,000 dead patients in 2018. It also indicates that the new patients with LC are gradually getting younger. According to the type of LC, it can be separated into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), in which NSCLC accounts for more than 85%.3 At present, the treatment of NSCLC is mainly through surgery combined with radiotherapy and chemotherapy. However, due to the relatively hidden incidence of NSCLC, most of the patients are in the middle and advanced stages when they are admitted to the hospital, while the prognosis of patients with advanced NSCLC is not ideal. Data show that the 5-year survival rate of patients with late NSCLC is less than 15%.4,5 In recent years, targeted therapy has played a vital role in improving the prognosis of patients.6 However, there are few therapeutic targets for NSCLC at present. It is the key to seek potential targets and further explore the molecular mechanism of NSCLC for improving the present situation.
At present, only 1% to 2% of the genome and transcriptome data studied by humans are genome encoded proteins, while most mammalian genomes contain this large amount of non-encoded RNA.7 In recent years, long-chain non-coding RNA (lncRNA) has become a research hotspot in various fields.8 LncRNA is a non-coding RNA with a length of more than 200 nt. Since lncRNA were initially found to have no protein-coding capacity,9 they were considered as metabolic wastes during transcription. However, recent studies have revealed that lncRNA has great differences in various diseases.10,11 Especially in tumors, researches12,13 have shown that part of lncRNA affect the development and progression of tumors. Nuclear para specific assembly transcript 1 (NEAT1) (also known as LINC00084), as a member of lncRNA family, is located on human 11q13.1 chromosome.14,15 Previous studies have revealed that NEAT1 is differentially expressed in liver carcinoma,16 gastric carcinoma17 and breast carcinoma.18 Many other studies have revealed that19,20 NEAT1 participates in the development of LC by regulating downstream microRNA (miR), but there is no report that NEAT1 can participate in the development of LC by regulating miR-204. In this study, we found a targeted binding site between NEAT1 and miR-204 through the joint prediction of LncBase,21 starbase,22 mircode23 and miRDB.24
Therefore, this study aimed to explore whether NEAT1 could regulate miR-204 and its downstream target genes to participate in the development of LC and provide potential targets for clinical treatment.
Methods and Materials
Online Website Analysis
The expression of miR-204 in lung adenocarcinoma (LA) was analyzed by starbase website (http://starbase.sysu.edu.cn/), and the expression of NEAT1 in LA was analyzed by GEPIA225 (http://gepia2.cancer-pku.cn/#index), and pictures were obtained to show the expression.
Clinical Data
A total of 68 patients with NSCLC treated in People’s Hospital of Gaochun from January 2015 to January 2017 were selected. The tumor and adjacent tissues of the patients were collected during surgery. The collected tissues were transported by liquid nitrogen and sent to the laboratory for testing. The samples collected from patients in this study had not been treated with chemoradiotherapy or anti-tumor therapy before this study. All samples of the patients were sent to the pathology department for examination and patients were confirmed as LA. All patients affixed informed consent forms. This research was ratified by the Medical Ethics Committee of People’s Hospital of Gaochun and it was consistent with the Helsinki Declaration.26
Cell Culturing and Transfection
Human bronchial epithelial cell strain 16HBE, as well as LC cell strains A549, SPC-A1, NCI-H23, NCI-H520 were acquired from the American Center for Typical Culture Collection (Manassas, Virginia, the United States). The purchased cells were cultivated using Dulbecco modified Eagle Medium (DMEM), which supplemented 10% of fetal bovine serum (Gibco, Grand Island, New York, the United States), penicillin (100 U/mL) and streptomycin (100 mg/mL) in a humid environment comprising 5% CO2 at 37 °C. In this study, Lipofectamine 3000 (Invitrogen, CA, USA) was used for transfection. miR-204 mimetic (miR-204-mimics)/inhibitor (miR-204-inhibit), siRNA targeting NEAT1 (si-NEAT1#1:5ʹ-GGTCTGTGTGGAAGGAGGAAGGCAG-3ʹ, si-NEAT1#2:5ʹ-GCCAUCAGCUUUGAAUAAAUU-3ʹ,si-NEAT1#3:5ʹ-GGUGUUAUCAAGUGAAUUAUU-3ʹ), lentivirus and corresponding negative control were all from GenePharma (Shanghai, China).
Detection of Cell Proliferation
In this study, CCK-8 kit was used to test cell proliferation. The transfected cells (2.5 × 10 3) were inoculated into 96-well plate (Corning Inc, United States). According to the manufacturer’s regulations, cell growth was evaluated at various time points (0, 24, 48, 72 and 96 h) through CCK-8 assay (Dojindo, Tokyo, Japan). The absorbance value at 450 nm wavelength was measured using an enzyme-labeling instrument (Molecular Devices, USA), which was positively correlated with cell proliferation.
Cell Invasion and Migration
In this study, Transwell was used to measure cell migration and invasion. For invasive analysis, the Transwell chamber (8 μm, Corning, Inc., Corning, NY, the United States) was precoated with Matrigel (BD Biosciences, Franklin Lake, NJ, the United States). Transfected cells were resuspended in serum-free medium and inoculated into the upper Transwell chamber. The DMEM (500 μL) comprising 10% fetal bovine serum was put in the lower chamber as chemotactic inducer. After culturing at 37 °C for 24 h, the cells remaining in the upper chamber were eliminated with cotton swabs, and the cells invading the lower chamber were fastened in 100% methanol and dyed with 0.05% crystal violet (Sigma-Aldrich). The amount of invasive cells was counted from 5 randomly selected areas and imaged using an inverted microscope (Olympus, Tokyo, Japan). The study of cell migration was conducted in a cross-well chamber without matrix and followed similar instructions.
Apoptosis Detection
Flow cytometry was used to test apoptosis in this study. Transfected cells were obtained by trypsin transfection. After double staining with FITC-Annexin V and PI, FITC-Annexin V apoptosis test kit (BD Biosciences) was used on the basis of kit instructions. Flow cytometer (FACScan, BD biosciences) of Cell Quest 3.0 software was used to analyze cells.
qRT-PCR
In this research, qRT-PCR was used to detect the relative expression of lncRNA/miR/mRNA. The cells were obtained and total RNA was abstracted from the tissue through TRIzol reagent (Invitrogen). The extracted total RNA was reverse transcribed into cDNA through RNA reverse transcription kit (Takara). Real-time PCR analysis was conducted with SYBR Green (Takara), the amplification was performed with ABI7500, and the operation was conducted on the basis of the manufacturer’s specifications. GAPDH (for lncRNAs) or U6 (for miRNAs) were used as internal parameters. The multiple change of relative expression was calculated by 2-−ΔΔ-CT.27 The primer sequences are shown in Table 1.
 |
Table 1 Primer Sequences |
WB Detection
Radioimmunoprecipitation assay (RIPA) of protease inhibitor was used to break cells with lysis buffer. After high-speed centrifugation, the supernatant was obtained and heated in a water bath to denature the protein. The protein was quantified by double caprylic acid (BCA). Then, the protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and sealed with 5% skim milk and 0.05%TBS-Tween-20 (TBST) for 1 h at room temperature, and the first antibodies (cleave-caspase 9, Bax, Bcl-2, MMP-2, MMP-9) were added and cultured at 4 °C for 12 h. The polyvinylidene fluoride (PVDF) film was washed with TBST solution, and then cultured with a secondary antibody (Hubeibiosci Biotechnology Co., Ltd., 1: 2000) at ambient temperature for 1 hour. After that, the chemiluminescence method was developed by hypersensitivity ECL (Hubei Biotechnology Co., Ltd.). β-actin was used as an internal reference.
Double Fluorescein Report
The miR-204 mutation site in NEAT1 luciferase reporter vector was produced through site-directed mutagenesis. PCR products were cloned into PMIR-GLO luciferase vector downstream of the firefly luciferase coding region (Invitrogen, USA). The construct was called pMIR-NEAT1-wt. The mutation of miR-204 binding site was introduced through site-directed mutagenesis, and the obtained plasmid was called pMIR-NEAT1-Mut. Luciferase assay was performed with 1×10 4, and HEK-293 cells were inoculated in a 48-hole plate. The co-transfection was conducted through 200 ng pMIR-NEAT1-wt or pMIR-NEAT1-Mut vector and 80 ng miR-204 mimetic or inhibitor. After transfection for 48 hours, the cells were obtained, and the contents of fireflies and algal luciferases were determined using Dual-Glo luciferase assay kit (Promega, Madison, Wisconsin, United States). All transfection tests were repeated three times. miR-204 and NUAK family kinase 1 (NUAK1) were carried out according to the above steps.
RIP Experiment
In this study, EZ-Magna RIP RNA binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA) was applied for RNA immunoprecipitation (RIP) analysis. The specific steps were carried out on the basis of the manufacturer’s kit. The method was as below: A549 cells with fusion degree of 80–90% were assimilated, obtained and lysed in RIP lysis buffer. Next, 100 μL of whole cell lysate was cultivated with RIP buffer comprising magnetic beads coupled to human antibody against Argonaut 2 (Ago2) (ABCAM, Cambridge, MA, USA) or normal mouse IgG (Millipore) for 6 hours at 4 °C. The beads were processed with protease K buffer to digest the protein, and then the target RNA was abstracted. Finally, the coprecipitated RNA was analyzed by qRT-PCR for further study.
Statistical Analysis
In this study, GraphPad 7 was used to draw the required pictures and data analysis. SPSS version 20.0 was used to analyze the independent prognostic factors of patients. The measurement data was represented by mean number ± standard deviation (Mean±SD). The comparison between groups was conducted through independent sample t test. The counting data were represented by percentage (%). The chi-square test was expressed by 2. One-way ANOVA was applied for comparison among multiple groups, expressed as F. LSD-t test was applied for pairwise comparison afterwards. Repetitive measurement and analysis of variance was applied for expression at multiple time points, expressed as F. Bonferroni was used for post test. Pearson test was used to analyze the correlation of each gene. There was a statistical difference with P<0.05.
Results
Expression of NEAT1 Increased in LA
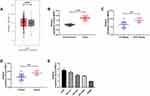
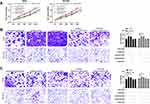
First of all, we analyzed it to determine the NEAT1 expression in LA through GEPIA2 online website. The results showed that the expression of NEAT1 in LA increased in the TCGA database (Figure 1A). Then, qRT-PCR was used to detect the expression in LA patients’ tissues and it revealed that the relative expression was obviously enhanced compared with the para-carcinoma tissues (Figure 1B). Patients were further separated into high and low expression groups in accordance with the median expression value of NEAT1. By observing the connection of NEAT1 with the pathological data of patients, it was found that there was lymph node metastasis and TNM high stage probability increased in patients with high expression of NEAT1 (Table 2). Further analysis uncovered that NEAT1 in patients with lymph node metastasis (Figure 1C) and high TNM stage (Figure 1D) was higher than in patients without lymph node metastasis and low TNM stage. In addition, we found that the NEAT1 in LC cell line was obviously higher than that in human bronchial epithelial cell line (Figure 1E) through qRT-PCR detection, suggesting that NEAT1 was highly expressed in LA.
 |
Table 2 Relationship Between NEAT1 and Pathological Data of LA Patients |
Knocking Down NEAT1 Could Inhibit the Growth, Invasion and Migration of LA Cells and Induce Cell Apoptosis
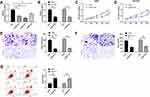
To further determine the influence of NEAT1 on LA, we constructed si-NEAT1 vector (Figure 2A), and selected si-NEAT1#2 with significant difference for cell transfection (Figure 2B). Subsequently, CCK-8 assay revealed that the proliferation activity of cells transfected with si-NEAT1#2 was significantly inhibited compared with that of si-NC (Figure 2C-D), while Transwell experiment revealed that the invasion and migration activity of cells transfected with si-NEAT1#2 were significantly decreased compared with that of si-NC (Figure 2E-F). In addition, flow cytometry showed that apoptosis was significantly induced after transfection of si-NEAT1#2 (Figure 2G). These results implied that NEAT1 might be a latent target for the therapy of LA.
NEAT1 Acted as an miR-204 Sponge
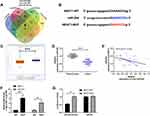
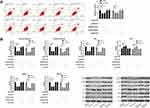
In order to further determine the mechanism of NEAT1 in LA, we predicted that NEAT1 could bind miR. Previous studies showed that lncRNA could regulate miR to participate in tumor development. Through the online prediction website, we predicted and analyzed the downstream of NEAT1 in combination with miR. It was found that NEAT1 and miR-204 had a targeted binding relationship (Figure 3A-B). Subsequently, we concluded that miR-204 was weakly expressed in LA through starbase analysis of miR-204 expression in LA (Figure 3C). In addition, qRT-PCR detection revealed that miR-204 in cancer tissue of LA patients was lower than that in para-carcinoma tissues (Figure 3D). The correlation analysis also revealed that the NEAT1 was negatively correlated with miR-204 in LA tissues (Figure 3E). This indicated that there was a latent targeting connection between NEAT1 and miR-204. Then, we found that miR-204-mimics could inhibit NEAT1-WT fluorescence activity (Figure 3F) through double luciferase report and RIP experiment analysis respectively, while RIP experiment showed that NEAT1 and miR-204 could be precipitated by Ago2 antibody (Figure 3G). It confirmed that NEAT1 could target miR-204.
miR-204 Targeted Regulation of NUAK1
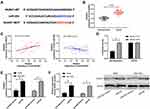
It has been proved that miR regulates downstream target genes to participate in the occurrence of diseases. In order to further study the potential mechanism of miR-204, we predicted the target gene downstream of miR-204. Through prediction, we found that there was a target site between miR-204 and NUAK1 (Figure 4A). Subsequently, we detected theNUAK1 in LA. qRT-PCR showed that the NUAK1 was increased in LA (Figure 4B). The correlation analysis also showed that miR-204 was negatively correlated with NUAK1 and positively correlated with NEAT1 in tumor tissue of patients with LA (Figure 4C). In addition, the double luciferase report experiment showed that the fluorescence activity of NUAK1-WT could be suppressed by miR-204-mimics (Figure 4D), and the relative expression of NUAK1 in the cells after transfection with miR-204-mimics was also determined through qRT-PCR and WB experiments (Figure 4E-F). This indicated that miR-204 targeted regulation of NUAK1.
NEAT1 Inhibited the Growth and Metastasis of LC Cells by Regulating miR-204/NUAK1 Axis
At the end of the study, we conducted a rescue experiment to further determine whether NEAT1 could participate in the development of LC by adjusting the miR-204/NUAK1 axis. Through the experiment, we concluded that the cell growth, invasion and migration abilities were significantly faster than those of pcDNA-NC+miR-inhibit, and the cell apoptosis was inhibited since miR-204-inhibit and pcDNA-NUAK1 were transfected into LA cells. However, the above results were reversed since si-NEAT1#2 was co-transfected with miR-204-inhibit or pcDNA-NUAK1 (Figure 5A-C, 6A). In addition, WB detection showed that the relative expression of cleaved-caspase 3, cleaved-caspase 9, Bax, MMP-2, MMP-9 protein in cells transfected with miR-204-inhibit and pcDNA-NUAK1 was obviously decreased, while the relative expression of Bcl-2 protein was obviously increased. However, the above results were reversed after co-transfection (Figure 6B), suggesting that NEAT1 inhibited the growth and metastasis of LC cells by regulating miR-204/NUAK1 axis.
Discussion
In this research, we concluded that NEAT1 expression increased in patients with LA and the cell strains, and the probability of TNM high staging and lymphatic metastasis increased significantly in patients with high expression of NEAT1. In addition, we found that knock-down of NEAT1 could inhibit the growth, invasion and migration of LA cells, and induce apoptosis of LA cells, so it was a potential therapy target for LA. Besides, we found for the first time that NEAT1 could suppress the growth and metastasis of LC through miR-204/NUAK1 axis, which was expected to become a clinical therapeutic target.
NEAT1, as an early lncRNA, has been found to affect the progression of many diseases. For instance, studies by Pang et al28 have revealed that NEAT1 could promote breast cancer progression through miR-124/STAT3 axis. In addition, the researches of Zhang et al29 have shown that NEAT1 can promote the progress of colorectal cancer by activating Wnt/β-catenin signal and interacting with DDX5. In this research, we first studied the NEAT1 in LA through GEPIA2 web analytics, and found that the NEAT1 in LA was higher than that in control samples. Then, we found that the expression of NEAT1 in tumor tissues of patients with LA and LA cell strains was higher than that in adjacent tissues and human bronchial epithelial cell lines through qRT-PCR detection, which was consistent with the previous research results, indicating that NEAT1 was expected to become a potential diagnostic indicator of LA. At present, the strategy for clinical treatment of tumors is to achieve therapeutic effect by suppressing the growth of tumor cells and facilitating tumor cell apoptosis.30 Therefore, we transfected the knocked-down NEAT1 vector into LA cells. Through observation, we found that the growth, invasion and migration of cells transfected with si-NEAT1#2 were significantly weakened compared with the control, and the apoptosis was further promoted. However, studies of Li et al31 have also shown that knocking down NEAT1 can suppress the proliferation and invasion of LC cells, which verified our study.
Competing endogenous RNAs (ceRNA) are considered as a new mechanism of RNA interaction.32,33 CeRNA can regulate gene expression by competitively binding microRNA.34 For example, researches by Yuan et al35 have shown that NEAT1/miR −133a axis participates in promoting the progression of cervical carcinoma by targeting SOX4. Other studies have revealed that36 NEAT1/miR-1224/KLF3 participates in proliferation, apoptosis and invasion of LC cells. In this study, we predicted the potential binding miR of NEAT1 through 4 online prediction websites and found that there was a targeted connection of NEAT1 with miR-204. Subsequently, we analyzed the miR-204 in LA through starbase and qRT-PCR. The results showed that the miR-204 in tumor samples was lower than that in control samples both in the database and in LA tumor tissues. The correlation analysis showed that there was a negative correlation of miR-204 with NEAT1. This indicated that there was a latent targeting connection of miR-204 with NEAT1. Then, we verified it by RIP and double luciferase reporting experiments. Previous studies have revealed that37 miR-204 is low in LC and is expected to become a potential diagnostic indicator for LC. Moreover, other studies have revealed that38 miR-204 participates in the development of LC through targeted regulation of downstream genes. NUAK1 is also known as adenosine monophosphate-related kinase 5 (ARK5). Studies have revealed that NUAK1 is differentially expressed in nasopharyngeal carcinoma39 and ovarian carcinoma,40 and it is expected to become a potential prognostic indicator. In this study, we found that there was a targeted binding site of miR-204 with NUAK1. The targeted relationship between miR-204 and NUAK1 has been confirmed in liver cancer in previous studies by Yu et al.41 However, it is not clear whether there is the same regulatory relationship in LC. In this research, we concluded that the NUAK1 in LA patients increased significantly and was negatively correlated with miR-204. In addition, we confirmed the targeting relationship between miR-204 and NUAK1 in LC through a double luciferase report.
More and more studies have revealed that lncRNA can participate in tumor growth by regulating the miR/mRNA axis.42 In the above studies, we found that NUAK1 might have the ability to regulate the miR-204/NUAK1 axis, but whether it can play a role is still unclear. Therefore, we conducted a rescue experiment at the end of our research. Through experiments, we found that the growth, invasion and migration of cells transfected with miR-204-inhibit and pcDNA-NUAK1 were significantly faster than those of the control, and the apoptosis of cells was inhibited. However, the above results were reversed since si-NEAT1#2 was co-transfected with miR-204-inhibit or pcDNA-NUAK1. In addition, our research also showed that the proteins of cleaved-caspase 3, cleaved-caspase 9, Bax, MMP-2 and MMP-9 in cells after co-transfection were obviously higher than those of cells transfected with miR-204-inhibit and pcDNA-NUAK1, while Bcl-2 was significantly lower, which suggested that NEAT1 could inhibit the growth of LA by regulating miR-204/NUAK1 axis to inhibit the invasion and migration of LA and promote the apoptosis of LA cells.
In this research, we confirmed the function of NEAT1/miR-204/NUAK1 axis in LA, but there are some limitations in this research. Firstly, we have not conducted in vivo experiments in this study, and whether NEAT1/miR-204/NUAK1 axis plays the same role in vivo still needs to be further verified. In addition, the samples that we collected in this study are single, and some reports have shown that lncRNA is expressed in various samples. Whether the development of LA can be judged by detecting other samples is also of great significance, so we hope to carry out more research in the future to further supplement our research results.
Conclusion
To sum up, the expression of NEAT1 increases in patients with LA and cell strains, and inhibits the growth of LA by mediating miR-204/NUAK1 axis. It is a potential therapeutic target for LA.
Funding
The authors received no funding for this work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Nasim F, Sabath BF, Eapen GA. Lung Cancer. Med Clin North Am. 2019;103(3):463–473. doi:10.1016/j.mcna.2018.12.006
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi:10.1038/nature25183
4. Rafei H, El-Bahesh E, Finianos A, Nassereddine S, Tabbara I. Immune-based therapies for non-small cell lung cancer. Anticancer Res. 2017;37(2):377–387. doi:10.21873/anticanres.11330
5. Reck M, Borghaei H, O’Byrne KJ. Nivolumab plus ipilimumab in non-small-cell lung cancer. Future Oncol. 2019;15(19):2287–2302. doi:10.2217/fon-2019-0031
6. Nagasaka M, Gadgeel SM. Role of chemotherapy and targeted therapy in early-stage non-small cell lung cancer. Expert Rev Anticancer Ther. 2018;18(1):63–70. doi:10.1080/14737140.2018.1409624
7. Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592(17):2884–2900. doi:10.1002/1873-3468.13182
8. Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74.
9. Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41(9):761–772. doi:10.1016/j.tibs.2016.07.003
10. Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705–1714. doi:10.1080/15476286.2017.1358347
11. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509.
12. Zhu L, Di Y, Liu G, Li C, Pan W, Li XJIJCEM. LncRNA CRNDE promotes hepatocellular carcinoma cell proliferation and upregulates cyclin D1 expression. 2018;11(7):6957–6964.
13. Mohammadrezakhani H, Baradaran B, Shanehbandi D, et al. Overexpression and clinicopathological correlation of long noncoding RNA TMPO-AS1 in colorectal cancer patients. J Gastrointest Cancer. 2019.
14. Li X, Wang S, Li Z, et al. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol. 2017;105(Pt 1):346–353. doi:10.1016/j.ijbiomac.2017.07.053
15. Wang L, Yang D, Tian R, Zhang H. NEAT1 promotes retinoblastoma progression via modulating miR-124. J Cell Biochem. 2019;120(9):15585–15593. doi:10.1002/jcb.28825
16. Li Y, Ding X, Xiu S, Du G, Liu Y. LncRNA NEAT1 promotes proliferation, migration and invasion via regulating miR-296-5p/CNN2 axis in hepatocellular carcinoma cells. Onco Targets Ther. 2019;12:9887–9897. doi:10.2147/OTT.S228917
17. Zhang J, Guo S, Piao HY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75(3):379–389. doi:10.1007/s13105-019-00690-8
18. Ke H, Zhao L, Feng X, et al. NEAT1 is required for survival of breast cancer cells through FUS and miR-548. Gene Regul Syst Bio. 2016;10(Suppl 1):11–17.
19. Chen Y, Qiu J, Chen B, et al. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-kappaB pathway. Int Immunopharmacol. 2018;59:252–260. doi:10.1016/j.intimp.2018.03.023
20. Jiang C, Zhong R, Zhang J, et al. Reduning injection ameliorates paraquat-induced acute lung injury by regulating AMPK/MAPK/NF-kappaB signaling. J Cell Biochem. 2019;120(8):12713–12723. doi:10.1002/jcb.28540
21. Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–238. doi:10.1093/nar/gkv1270
22. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–97. doi:10.1093/nar/gkt1248
23. Xia T, Liao Q, Jiang X, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi:10.1038/srep06088
24. Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18. doi:10.1186/s13059-019-1629-z
25. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
26. Issue Information-Declaration of Helsinki. J Bone Miner Res. 2018;33(12):BM i–BM ii. doi:10.1002/jbmr.3259
27. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25(4):402–408. doi:10.1006/meth.2001.1262
28. Pang Y, Wu J, Li X, et al. NEAT1/miR124/STAT3 feedback loop promotes breast cancer progression. Int J Oncol. 2019;55(3):745–754.
29. Zhang M, Weng W, Zhang Q, et al. The lncRNA NEAT1 activates Wnt/beta-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11(1):113. doi:10.1186/s13045-018-0656-7
30. Goldar S, Khaniani MS, Derakhshan SM, Baradaran B. Molecular mechanisms of apoptosis and roles in cancer development and treatment. Asian Pac J Cancer Prev. 2015;16(6):2129–2144. doi:10.7314/APJCP.2015.16.6.2129
31. Li S, Yang J, Xia Y, Fan Q, Yang KP. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res. 2018;26(2):289–296. doi:10.3727/096504017X15009404458675
32. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
33. Liu H, Xu D, Zhong X, et al. LncRNA-mRNA competing endogenous RNA network depicts transcriptional regulation in ischaemia reperfusion injury. J Cell Mol Med. 2019;23(3):2272–2276. doi:10.1111/jcmm.14163
34. Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148–64167. doi:10.18632/oncotarget.11637
35. Yuan LY, Zhou M, Lv H, et al. Involvement of NEAT1/miR-133a axis in promoting cervical cancer progression via targeting SOX4. J Cell Physiol. 2019;234(10):18985–18993. doi:10.1002/jcp.28538
36. Yu PF, Wang Y, Lv W, et al. LncRNA NEAT1/miR-1224/KLF3 contributes to cell proliferation, apoptosis and invasion in lung cancer. Eur Rev Med Pharmacol Sci. 2019;23(19):8403–8410.
37. Guo W, Zhang Y, Zhang Y, et al. Decreased expression of miR-204 in plasma is associated with a poor prognosis in patients with non-small cell lung cancer. Int J Mol Med. 2015;36(6):1720–1726. doi:10.3892/ijmm.2015.2388
38. Wang P, Lv HY, Zhou DM, Zhang EN. miR-204 suppresses non-small-cell lung carcinoma (NSCLC) invasion and migration by targeting JAK2. Genet Mol Res. 2016;15:2.
39. Liu J, Tang G, Huang H, Li H, Zhang P, Xu L. Expression level of NUAK1 in human nasopharyngeal carcinoma and its prognostic significance. Eur Arch Otorhinolaryngol. 2018;275(10):2563–2573. doi:10.1007/s00405-018-5095-0
40. Phippen NT, Bateman NW, Wang G, et al. NUAK1 (ARK5) is associated with poor prognosis in ovarian cancer. Front Oncol. 2016;6:213. doi:10.3389/fonc.2016.00213
41. Yu Y, Wang Y, Xiao X, et al. MiR-204 inhibits hepatocellular cancer drug resistance and metastasis through targeting NUAK1. Biochem Cell Biol. 2019;97(5):563–570. doi:10.1139/bcb-2018-0354
42. Liu HY, Lu SR, Guo ZH, et al. lncRNA SLC16A1-AS1 as a novel prognostic biomarker in non-small cell lung cancer. J Investig Med. 2020;68(1):52–59. doi:10.1136/jim-2019-001080
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.