Back to Journals » Cancer Management and Research » Volume 13
LncRNA NEAT1 Acts as an miR-148b-3p Sponge to Regulate ROCK1 Inhibition of Retinoblastoma Growth
Authors Lu H, Zhang Z, Lu Y, Xiu WW, Cui J
Received 8 July 2020
Accepted for publication 15 October 2020
Published 12 July 2021 Volume 2021:13 Pages 5587—5597
DOI https://doi.org/10.2147/CMAR.S271326
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Hang Lu,1,* Zhenjun Zhang,2,* Yao Lu,3 Weiwei Xiu,1 Jinglin Cui1
1Research Center of Ophthalmology, The First Hospital of Qiqihar, Affiliated Qiqihar Hospital, Southern Medical University, Qiqihar, Heilongjiang Province, People’s Republic of China; 2Ophthalmology Department, Beiman Hongpeng Hospital of Qiqihar, Qiqihar, Heilongjiang Province, People’s Republic of China; 3International Education College, Heilongjiang University of Chinese Medicine, Harbin, Heilongjiang Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jinglin Cui
Research Center of Ophthalmology, The First Hospital of Qiqihar, Affiliated Qiqihar Hospital, Southern Medical University, No. 30 Gongyuan Road, Longsha District, Qiqihar, Heilongjiang Province, People’s Republic of China
Email [email protected]
Background: It is reported that long non-coding RNA nuclear paraspeckle assembly transcript 1 (LncRNA NEAT1) is involved in the occurrence and development of various cancers. However, the detailed biological function and mechanism of LncRNA NEAT1 in retinoblastoma are still unclear. So we will explore the biological function and possible mechanism of LncRNA NEAT1 in retinoblastoma.
Materials and Methods: Quantitative real-time PCR (qRT-PCR) was used to detect LncRNA NEAT1 in retinoblastoma tissues and cell lines. Cell counting kit 8, Transwell and flow cytometry were applied to explore cell proliferation, invasion and apoptosis. The target miRNAs (miR) of LncRNA NEAT1 and miR and downstream target genes were predicted using Starbase3.0 software and confirmed by double luciferase reporting test and RNA binding protein immunoprecipitation (RIP). Western Blot was applied to explore ROCK1 in cells, and tumor allogeneic experiment was applied to study the role of LncRNA NEAT1 on tumor growth.
Results: It was found that LncRNA NEAT1 was up-regulated in retinoblastoma tissues, cells and serum, and the prognosis of patients with high expression of LNC RNA NEAT 1 was poor. Functional analysis showed that knocking down LncRNA NEAT1 could weaken proliferation and invasion, and accelerate apoptosis. Tumor allogeneic experiment showed that sh-NEAT1 injection can inhibit tumor growth. In addition, LncRNA NEAT1 inhibited proliferation and invasion, and promoted apoptosis through miR-148b-3p/ROCK1 axis.
Conclusion: LncRNA NEAT1 can mediate miR-148b-3p/ROCK1 axis to weaken the proliferation and invasion of retinoblastoma.
Keywords: LncRNA NEAT1, miR-148b-3p, ROCK1, retinoblastoma, proliferation, invasion
Core Tips
LncRNA NEAT1 is up-regulated in retinoblastoma, and the prognosis of high expression is poor. Knocking down LncRNA NEAT1 can reduce the proliferation, invasion, and apoptosis through miR-148b-3p/ROCK1 axis, which is a potential target for retinoblastoma treatment.
Introduction
Retinoblastoma is a clinically common intraocular malignant tumor, which is mainly developed from immature cells in retina and is more common in children under 5 years old.1,2 Data show that3 one retinoblastoma occurs in every 14,000–20,000 newborns, accounting for 2–4% of malignant tumors in children. Although surgery combined with radiotherapy and chemotherapy has made some progress, the therapeutic effect and prognosis of retinoblastoma are still unsatisfactory.4,5 We believe that this is mainly due to delayed diagnosis and high incidence of tumor metastasis.6,7 Therefore, we urgently need to explore the relevant mechanisms of retinoblastoma and provide effective therapeutic schemes.
LncRNA is a non-coding RNA, which is more than 200nt.8 Previous studies showed that9 LncRNA is unable to encode proteins and is considered by scholars as a noise generated during transcription. However, as more and more studies have found that LncRNA is unable to directly encode proteins, LncRNA participates in gene expression regulation at transcription and post-transcription levels. Through regulating the interaction among DNA, protein and RNA, LncRNA (Human body) drives the biological functions of various cancer cells (Human Tumor) to change and plays a role in promoting tumor growth.10,11 Nuclear paraspeckle assembly transcript 1 (LncRNA NEAT1) is over-expressed in a variety of tumors (Human Tumor),12,13 and studies have confirmed that LncRNA NEAT1 is participated in a number of processes (LncRNA NEAT1 is involved in many processes in human body), including cell differentiation, inflammation and stress response.14,15 However, there are very few studies on the relationship between LncRNA NEAT1 and retinoblastoma. Previous studies have found that16 LncRNA NEAT1 is highly expressed in retinoblastoma (Human retinoblastoma). The specific mechanism is still unclear.
Competing endogenous RNAs (ceRNA) are used to describe the relationship between LncRNA and microRNA (miR), that is, LncRNA silences miR by competing with microRNA response elements (MREs) and miR elements to inhibit miR expression, which indirectly affects miR target gene transcription and expression.17 Through online prediction, we found that there was a targeted binding site between NEAT1 and miR-148b-3p, and in previous studies, it was found that miR-148b-3p was low in retinoblastoma, indicating that there may be a potential targeted regulatory relationship between NEAT1 and miR-148b-3p.
Therefore, in order to confirm our conjecture, this study provided reference for clinical treatment and diagnosis by exploring the relevant mechanism of LncRNA NEAT1 in retinoblastoma.
Materials and Methods
Data Collection
Altogether 50 children with retinoblastoma treated in our hospital from May 2012 to March 2014 were collected as the patient group, and 30 healthy children were collected as the control group. Inclusion criteria for the children: the children were diagnosed as retinoblastoma by pathological biopsy; the children met the 7th edition of TNM staging system issued by American Joint Committee on Cancer (AJCC); all the children were monocular tumors; the guardian of the children was signed an informed consent form. Exclusion criteria for the children: children had congenital defects and were complicated with other tumors or infections; children have received anti-tumor treatment before this study and cannot be completely followed-up. The follow-up of children was counted by telephone and electronic pathology files in outpatient department. Follow-up was conducted in the 1st, 3rd, 6th, 9th and 12th month of each year, for a total of 5 years. This study was approved by the Medical Ethics Committee of The First Hospital of Qiqihar. This study fully conformed to the Declaration of Helsinki.
Sample Collection
Before treatment, 5mL of peripheral blood was collected from patients and normal children, placed for 30min, centrifuged at 1500× g at 24°C for 10min, and then the serum was collected. In addition, tumor tissues of children were collected as samples for this study, and 22 normal retinal tissue samples (normal retinal tissue samples were all from ruptured spheres) were collected as controls.
Cell Culture
Retinoblastoma Y79, WERI-Rb1 and SO-RB-50 cell lines and normal human epithelial retinal cell ARPE-19 were purchased from ATCC (Manacas, Virginia, USA). The repurchased cells were transferred to RPMI 1640 medium for culture, containing 10% bovine fetal serum (FBS, Gibco, USA), and the cells were cultured in 5% CO2 at 37°C.
Cell Transfection
pcDNA was used as sequence vector, three LncRNA NEAT1 small interfering RNA (siRNA, si-NEAT1#1, #2, #3) and negative control sequences (si-NC) were designed and synthesized according to GenePharma. In addition, pcDNA was used to construct rho-associated coiled-coil-containing protein kinase 1 (ROCK1) over-expression vector (pcDNA-ROCK1) and interference sequence (si-ROCK1). In addition, the above primer sequences for constructing miR-148b-3p inhibitor, miR-148b-3p-mimics and blank control (miR-NC) were designed and synthesized by Sangon Biotech Co., Ltd. Lipofectamine 2000 (Invitrogen, Pittsburgh, PA, USA) was used, the above substances were transferred into Y79, WERI-Rb1 cells, and the cells were obtained 48 hours after transfection, and the transfection efficiency was detected by quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
qRT-PCR Detection
Total RNA was obtained from the samples by TRIzol kit (Invitrogen, USA), and the purity, concentration and integrity were detected by UV spectrophotometer and agarose gel electrophoresis. Then, total RNA was reverse transcribed by TaqMan™ Reverse Transcription Kit (Invitrogen, USA), and the transcription steps were operated according to the kit instructions. Quantitative detection was carried out. The 7500 Fast Dx Real-Time PCR Instrument (Applied Biosystems™, USA) was applied. PCR reaction system: 12.5μ L of SYBR® Premix Ex TapTM II (2X), 1μL of upstream and downstream primers, 2μL of Templat *2, 8.5μL of dH2O, with a total of the above of 25μL. PCR reaction conditions: pre-denaturation at 95°C for 30s, denaturation at 95°C for 5s, annealing and extension at 60°C for 30s, with a total of 40 cycles. In the experiment, three parallel repeating wells were designed, and all specimens were repeatedly tested for 3 times. U6 and GADPH were applied as internal parameters, and 2−Δct was used to analyze the data.18
Western Blot Analysis
The collected cells were cracked with RIPA buffer (Cell Signal Technology, Inc., MA, USA), protein concentration and quantification were determined with BCA kit (Beyotime Biotechnology, Shanghai, China), electrophoretic separation was performed with 10%SDS gel, and the cells were transferred to PVDF (EMD Milliperate Corporation, Billerica, MA, USA). Altogether 5% defatted milk was used for sealing, primary anti-ROCK 1 (1:1000) was added. β-action (1:1000) was used as internal reference, incubated at 4°C for a night. The membrane was then rinsed with TBST (5 min × 3). Goat anti-mouse/rabbit IgG coupled with horseradish peroxidase (R&D Company, USA) was added, cultivated at room temperature for 2h. Finally, the print was detected with ECL chemiluminescence reagent (Thermo Fisher Scientific, USA) and Amersham Prime ECL Plus was used for detection. The experiment was repeated six times.
Double Fluorescein Report
Starbase3.0 was applied to find the potential binding site of NEAT with miR-148b-3p, and Starbase3.0, TargetScan and miRDB were applied to find the targeted binding site of miR-148b-3p with ROCK1. Complementary DNA fragments containing wild-type (NEAT-WT) or mutant NEAT (NEAT-MUT) fragments were sub-cloned to the downstream of the luciferase gene in psi-CHECK2. In addition, ROCK1 (ROCK1-WT) and corresponding mutant (ROCK1-MUT) fragments were established, and miR-148b-3p simulation or miR-148b-3p inhibitor were co-transfected with NEAT/ROCK1 report vector (Invitrogen, USA) into Y79 cells by transfection reagent. Fireflies and renin luciferase activities in cell lysates were continuously measured using a dual-luciferase reporting kit (Promega, USA) after 48h of transfection.
Immunocoprecipitation (RIP)
Magna RIP RNA binding protein immunoprecipitation kit (MILLIALE, Billerica, MA) was applied for operation, and the operation steps were carried out according to the kit instructions. The specific steps were as follows: RB cells were cracked in RIP lysis buffer. Then cell lysate, RIP buffer including magnetic beads with human anti-Ago2 antibody and normal mouse IgG (as negative control) were incubated at 4°C for 4 hours, immunoprecipitation RNA was extracted and qRT-PCR analysis was performed.
Tumor Allogeneic Experiment
Male BALB/c nude mice (4 weeks old) were purchased and transferred to the Animal Experimental Center. In the room without specific pathogen (constant temperature: 22°C-25°C, constant humidity: 40%-50%), Y79 cells were adjusted to 1× 106 cells and suspended in 100μL PBS. Then, the cells were injected into two sides of nude mice, cultured for 4 weeks. The tumor was detected by vernier caliper every Sunday, and the tumor volume was measured. The calculation formula was as follows: V = 0.5* width2* length. The tumor tissue of nude mice was collected after 4 weeks to test tumor mass and the expression in tumor tissue. This study was approved by the Animal Ethical Protection Association of Qiqihar First Hospital. In addition, the experiment was conducted in accordance with NIH guidelines for the care and use of laboratory animals.
Cell Proliferation Detection
The transfected cells were collected, adjusted to 1×103 cells/well, transferred to a 96-well plate, and 10μL of CCK-8 solution was added to each detection point. After incubation for 2 hours, the cells were detected by using an enzyme-labeled instrument at 450mm absorbance. The experiment was repeated six times.
Cell Invasion Detection
Detection was performed using Transwell technology (Corning, USA) by adjusting the number of cells to 5*104 and inoculating to the upper chamber. Then, 750μL FBS was put in the lower chamber. The invaded cells were fixed with 4% paraformaldehyde for 30min. After washing with PBS, the cells were stained with 0.1% crystal violet for 15min and counted with a microscope. The experiment was repeated six times.
Apoptosis Detection
Flow cytometry was used to detect apoptosis and cell cycle. The specific steps were as follows. After 48 hours of cell transfection, the cell suspension was adjusted to 1*106 cells and inoculated into cell culture flasks for overnight growth. The collected cells were rinsed with PBS, and the apoptosis rate was measured by Annexin V-FITC apoptosis detection kit (Invitrogen™, USA). The detection steps were carried out according to the kit instructions. The experiment was repeated six times.
Statistical Analysis
GraphPad 7 was applied to visualize required pictures and to conduct data analysis. SPSS20.0 was used to analyze independent prognostic factors of patients, and K–S test to analyze dose data distribution. The normal distribution data were expressed by mean± standard deviation (Meas±SD), and independent sample t test was applied for inter-group comparison. The counting data were expressed by percentage (%), chi-square test was expressed by χ2. One-way ANOVA was used for comparison among multiple groups, which was expressed by F. LSD-t test was applied for post-event comparison, repeated measurement ANOVA was used for expression at multiple time points, which was expressed by F. Bonferroni was used for back testing. The diagnostic value of Lnc NEAT in retinoblastoma was analyzed by ROC curve. Pearson test was used to analyze the correlation of each gene, K-M survival curve was used to visualize the total survival condition of patients, Log rank test was applied for analysis. When P < 0.05, there was statistical difference.
Results
High Expression of LncRNA NEAT1 in Retinoblastoma Patients
We detected the tissues and serum to verify the expression of LncRNA NEAT1. The results showed that retinoblastoma was up-regulated (Figure 1A-B, P<0.001), and LncRNA NEAT1 in tissue and serum was positively related through Pearson correlation analysis (Figure 1C, R=0.177, P=0.002). Moreover, the patients were grouped into high and low expression group according to the median value of LncRNA NEAT1. By analyzing the correlation of LncRNA NEAT1 with pathological data of patients, it was showed that LncRNA NEAT1 increased, tumor diameter≥10cm (P=0.021), TNM high staging (P=0.004), and the probability of low differentiation increased (P=0.004) significantly (Table 1). We further visualized ROC curve according to the expression of LncRNA NEAT1 to determine the diagnostic value of LncRNA NEAT1 in retinoblastoma, and found that the area under LncRNA NEAT1 curve was 0.891 (Figure 1D). In addition, we visualized 5-year survival curve through follow-up and found that the 5-year survival rate of high expression group was lower than that low expression group (Figure 1E). Cox regression analysis indicated that LncRNA NEAT1 was an independent prognostic factor (Table 2).
 |
Table 1 Relationship Between LncRNA NEAT1 and Pathological Data |
 |
Table 2 Cox Regression Analysis |
Knocking Down LncRNA NEAT1 Can Inhibit Retinoblastoma Growth
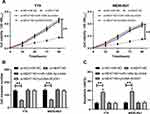
The role of LncRNA NEAT1 was determined through the above research. In order to further explore its related mechanism, we further carried out cell experiments. First, we found that the expression of LncRNA NEAT1 in retinoblastoma cells increased significantly (Figure 2A, P<0.05). Then, we established three si-NEAT1 sequences. Further tests found that the inhibition effect of si-NEAT1#2 plasmid was the most significant (Figure 2B, P<0.01). After transfection, it was showed that LncRNA NEAT1 in cells decreased obviously (Figure 2C, P<0.01). CCK-8 experiment and Transwell experiment found that the proliferation and invasion ability of LncRNA NEAT1 knock-down cells were significantly weakened compared with si-NC (Figure 2D-E, P<0.01), while flow cytometry showed that the apoptosis rate of LncRNA NEAT1 knock-down cells was obviously increased compared with si-NC (Figure 2F, P<0.05). In addition, nude mice experiment found that the tumor volume and mass of nude mice were significantly reduced after stable injection of sh-NEAT1 (Figure 2G-I, P<0.01).
LncRNA NEAT1 Can Be Used as miR-148b-3p Sponge
We predicted the potential miR of LncRNA NEAT1 to further find the relevant mechanism and found that there were binding sites of miR-148b-3p with LncRNA NEAT1 (Figure 3B). In order to verify the expression of miR-148b-3p in retinoblastoma, we analyzed GSE41321 chip through GEO database online geo2R software and concluded that miR-148b-3p was down-regulated in retinoblastoma (Figure 3A, P<0.001). In addition, we also carried out double luciferase activity detection and RIP experiment to confirm. Double luciferase activity detection found that NEAT1-WT could be reduced by miR-148b-3p-mimics, while NEAT1-MUT fluorescence activity was not affected (Figure 3C, P<0.01). RIP experiment found that Ago2 antibody could precipitate NEAT1 and miR-148b-3p, but IgG antibody could not precipitate (Figure 3D, P<0.01). After transfection of miR-148-3p-mimics/inhibit into cells, it was found that miR-148-3p was obviously over-expressed and inhibited (Figure 3E, P<0.01). Cell experiments indicated that up-regulation of miR-148b-3p can inhibit proliferation (Figure 3F, P<0.01) and invasion (Figure 3G, P<0.01) of retinoblastoma and promote the increase of apoptosis rate (Figure 3H, P<0.05).
miR-148b-3p Targeting ROCK1 to Inhibit Cell Growth
The main mechanism of miR was to regulate downstream target genes so as to participate in biological processes. The potential target genes of miR-148b-3p were analyzed through online prediction software to further find the mechanism, and binding sites of ROCK1 with miR-148b-3p were found through combined prediction through three online prediction websites (Figure 4A). In order to further verify the relationship between the two, we found through double luciferase detection that miR-148b-3p-mimics reduce the fluorescence activity of ROCK1-WT, and the expression of ROCK1 in cells was reduced after transfection of miR-148b-3p-mimics (Figure 4B-C, P<0.01). Cell experiments showed that the proliferation (Figure 4H-I, P<0.01) and invasion ability of cells transfected with si-ROCK1 reduced, and the apoptosis rate (Figure 4J, P<0.01) was up-regulated. However, the proliferation, invasion and apoptosis of cells transfected with miR-148b-3p-inhibit and si-ROCK1 had no difference compared with si-NC (Figure 4D-G, P<0.01).
LncRNA NEAT1 Acts as miR-148b-3p Sponge to Regulate ROCK1 to Inhibit Cell Proliferation and Invasion
We co-transfected plasmids to verify the relationship of LncRNA NEAT1 with miR-148b-3p/ROCK1 axis, and established si-NEAT1#2+pcDNA-ROCK1, si-NEAT1#2+miR-148b-3p-inhibit, si-NEAT1#2, si-NC+miR-NC, respectively, to further observe cell growth. The results showed that CCK-8 (Figure 5A, P<0.01), Transwell (Figure 5B, P<0.01) and Flow cytometry (Figure 5C, P<0.01) experiment found that the co-transfection of pcDNA-ROCK1, miR-148b-3p-inhibit and si-NEAT1#2 could reverse the effect of knock-down NEAT1.
Discussion
In this research, we confirmed that LncRNA NEAT1 was up-regulated in retinoblastoma, and the prognosis of patients with high expression was poor. In addition, we further found through basic research that knock-down LncRNA NEAT1 could inhibit proliferation, invasion, and apoptosis of retinoblastoma cells through miR-148b-3p/ROCK1 axis, which may be a potential target for retinoblastoma treatment.
LncRNA, as a long-chain non-coding RNA, was previously considered by scholars as “noise” generated during transcription due to non-coding protein.19 However, in recent years, more and more researches have found that LcnRNA can be used as ceRNA of miR and regulate miR expression through competing MREs.20 LncRNA NEAT1 is LncRNA located on human chromosome 14q32. Previous studies have found that LncRNA NEAT1 has a high expression in gastric cancer, liver cancer and lung cancer.21–23 However, it was rarely reported on the effect of LncRNA NEAT1 in retinoblastoma. Wang et al24 showed that LncRNA NEAT1 increased in retinoblastoma and was expected to become a potential therapeutic target for retinoblastoma. However, the relevant mechanism has not been further elaborated. Therefore, we conducted further research in this study to verify the relevant role of LncRNA NEAT1 in retinoblastoma.
We first detected LncRNA NEAT1 in retinoblastoma. Through detection, it was showed that LncRNA NEAT1 in both tissues and serum was up-regulated, and the detection of LncRNA NEAT1 in serum could be used as a potential diagnostic indicator for retinoblastoma. Correlation analysis found that the expression of LncRNA NEAT1 in tissues and serum was positively correlated. This indicated that LncRNA NEAT1 is also stably expressed in serum. Further analysis found that the probability of TNM high and low differentiation in patients with LncRNA NEAT1 over-expression increased. In addition, survival follow-up found that the 5-year survival rate of patients with LncRNA NEAT1 over-expression down-regulated. Cox regression analysis indicated that LncRNA NEAT1 could be used as an independent prognostic factor for retinoblastoma. However, we found through basic research that knocking down LncRNA NEAT1 inhibited the growth of retinoblastoma.
ceRNA is an important mechanism for LncRNA to participate in biological processes in recent years and has been accepted by many scholars.25 For example, research by An et al26 found that LncRNA NEAT1 regulates the expression of ZEB1 through miR-194 to participate in the drug resistance of ovarian cancer cells to paclitaxel, and other research showed that27 LncRNA NEAT1 promotes the occurrence of laryngeal squamous cell carcinoma. In addition, we found a binding target of miR-148b-3p with LncRNA NEAT1 through online prediction. miR-148b-3p is down-regulated in various tumors and is a tumor suppressor gene.28–30 However, there has been no previous study on miR-148b-3p and retinoblastoma. For this reason, we first found that miR-148b-3p is down-regulated in retinoblastoma through online analysis of GEO database. Then, we confirmed that there is indeed a targeted relationship of the two through double luciferase report and RIP experiment. Through experiments, it was showed that cell proliferation and invasion ability were inhibited after over-expressing miR-148b-3p, while cell apoptosis was increased, and cell biological function was reversed after knocking down miR-148b-3p.
miR regulates downstream target genes to change the expression of target genes, which is a classical pathway of miR.31 We further predicted through Starbase3.0, TargetScan, and miRDB to determine the role of miR-148b-3p, and found that ROCK1 and miR-148b-3p have potential binding sites. ROCK1 is a protein serine/threonine kinase located on chromosome 18q11.1, which takes effect on tumor invasion.32 It was further concluded that miR-148b-3p-mimics can inhibit the fluorescence activity of ROCK1-WT through double luciferase report, and the expression of both ROCK1 protein and mRNA is obviously inhibited after cell transfection miR-148b-3p-mimics/inhibit. Cell experiments also found that the cell proliferation and invasion ability were reduced and the apoptosis rate increased after knocking down ROCK1, while the results were reversed after co-transfection of miR-148b-3p-inhibit and si-ROCK1. In order to verify the correlation of LncRNA NEAT1 with miR-148b-3p/ROCK1 axis, we conducted co-transfection and further detected the cell growth. CCK-8 experiment indicated that the co-transfection of pcDNA-ROCK1, miR-148b-3p-inhibit and si-NEAT1#2 could reverse the knock-down of NEAT1 on cell proliferation, invasion and apoptosis.
Through the above research, we could basically confirm that LncRNA NEAT1 can act as miR-148b-3p sponge to regulate ROCK1 to inhibit the growth of retinoblastoma. However, there are still some limitations. Firstly, the expression of miR-148b-3p and ROCK1 in patient tissues was not explored in this study. Secondly, the samples in this study are relatively small. Finally, it is unclear whether LncRNA NEAT1 can participate in the progress of diseases through other mechanisms in retinoblastoma. Therefore, we hope to collect more samples in future research and further explore the possible pathway of LncRNA NEAT1 in retinoblastoma through bioinformatics analysis, so as to improve our research conclusion.
To sum up, LncRNA NEAT1 can mediate miR-148b-3p/ROCK1 axis to inhibit proliferation and invasion of retinoblastoma.
Acknowledgment
These authors contributed equally to this study as co-first authors: Hang Lu and Zhenjun Zhang.
Funding
This study was supported by Heilongjiang Natural Science Foundation Joint Guidance Project. Project Number: LH2019H063
Disclosure
The authors report no conflicts of interest for this work.
References
1. Kivela T The epidemiological challenge of the most frequent eye cancer: retinoblastoma, an issue of birth and death. Br J Ophthalmol. 2009;93(9):1129–1131. doi:10.1136/bjo.2008.150292
2. Broaddus E, Topham A, Singh AD Incidence of retinoblastoma in the USA: 1975-2004. Br J Ophthalmol. 2009;93(1):21–23. doi:10.1136/bjo.2008.138750
3. Wei D, Miao Y, Yu L, Wang D, Wang Y Downregulation of microRNA198 suppresses cell proliferation and invasion in retinoblastoma by directly targeting PTEN. Mol Med Rep. 2018;18(1):595–602.
4. Damodaran S, Bajaj MS, Sharma P, et al. Swept-source optical coherence tomography features of regressed macular retinoblastoma. Indian J Ophthalmol. 2019;67(12):2013–2018. doi:10.4103/ijo.IJO_533_19
5. Kabre RS, Kamble KM Retinoblastoma: a retrospective analysis of 141 patients from 1983 to 2013 at a tertiary care hospital in Nagpur, India. South Asian J Cancer. 2019;8(3):195–197. doi:10.4103/sajc.sajc_314_18
6. Kaliki S, Gupta S, Ramappa G, Mohamed A, Mishra DK High-risk retinoblastoma based on age at primary enucleation: a study of 616 eyes. Eye (Lond). 2020;34(8):1441–1448. doi:10.1038/s41433-019-0698-2
7. Li Z, Guo J, Xu X, Wang Y, Mukherji SK, Xian J Diagnosis of postlaminar optic nerve invasion in retinoblastoma with MRI features. J Magn Reson Imaging. 2020;51(4):1045–1052. doi:10.1002/jmri.26961
8. Ishola AA, La’ah AS, Le HD, et al. Non-coding RNA and lung cancer progression. J Chin Med Assoc. 2020;83(1):8–14. doi:10.1097/JCMA.0000000000000225
9. Mohammadrezakhani H, Baradaran B, Shanehbandi D, et al. Overexpression and clinicopathological correlation of long noncoding RNA TMPO-AS1 in colorectal cancer patients. J Gastrointest Cancer. 2020;51(3):952–956. doi:10.1007/s12029-019-00333-7
10. Tu C, Ren X, He J, et al. The value of LncRNA BCAR4 as a prognostic biomarker on clinical outcomes in human cancers. J Cancer. 2019;10(24):5992–6002. doi:10.7150/jca.35113
11. Yu H, Xu Y, Zhang D, Liu G Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun. 2018;503(3):2095–2100. doi:10.1016/j.bbrc.2018.07.165
12. Li Y, Chen D, Gao X, Li X, Shi G LncRNA NEAT1 regulates cell viability and invasion in esophageal squamous cell carcinoma through the miR-129/CTBP2 axis. Dis Markers. 2017;2017:5314649. doi:10.1155/2017/5314649
13. Li X, Wang S, Li Z, et al. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol. 2017;105Pt1: 346–353. doi:10.1016/j.ijbiomac.2017.07.053
14. Zhang P, Cao L, Zhou R, Yang X, Wu M The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;10(1):1495. doi:10.1038/s41467-019-09482-6
15. Huang-Fu N, Cheng JS, Wang Y, Li ZW, Wang SH Neat1 regulates oxidized low-density lipoprotein-induced inflammation and lipid uptake in macrophages via paraspeckle formation. Mol Med Rep. 2018;17(2):3092–3098.
16. Zhong W, Yang J, Li M, Li L, Li A Long noncoding RNA NEAT1 promotes the growth of human retinoblastoma cells via regulation of miR-204/CXCR4 axis. J Cell Physiol. 2019;234(7):11567–11576. doi:10.1002/jcp.27812
17. Bossi L, Figueroa-Bossi N Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat Rev Microbiol. 2016;14(12):775–784. doi:10.1038/nrmicro.2016.129
18. Livak KJ, Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego, Calif). 2001;25(4):402–408. doi:10.1006/meth.2001.1262
19. Ferre F, Colantoni A, Helmer-Citterich M Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–116. doi:10.1093/bib/bbv031
20. Yu HM, Wang C, Yuan Z, Chen GL, Ye T, Yang BW LncRNA NEAT1 promotes the tumorigenesis of colorectal cancer by sponging miR-193a-3p. Cell Prolif. 2019;52(1):e12526. doi:10.1111/cpr.12526
21. Qi L, Liu F, Zhang F, et al. lncRNA NEAT1 competes against let-7a to contribute to non-small cell lung cancer proliferation and metastasis. Biomed Pharmacother/Biomedicine & Pharmacotherapie. 2018;103:1507–1515. doi:10.1016/j.biopha.2018.04.053
22. Fu JW, Kong Y, Sun X Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142(7):1571–1579. doi:10.1007/s00432-016-2152-1
23. Guo S, Chen W, Luo Y, et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8(5):5395–5402.
24. Wang L, Yang D, Tian R, Zhang H NEAT1 promotes retinoblastoma progression via modulating miR-124. J Cell Biochem. 2019;120(9):15585–15593. doi:10.1002/jcb.28825
25. Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148–64167. doi:10.18632/oncotarget.11637
26. An J, Lv W, Zhang Y LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. Onco Targets Ther. 2017;10:5377–5390. doi:10.2147/OTT.S147586
27. Wang P, Wu T, Zhou H, et al. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J Exp Clin Cancer Res. 2016;35:22. doi:10.1186/s13046-016-0297-z
28. Wang Y, Li J, Kuang D, et al. miR-148b-3p functions as a tumor suppressor in GISTs by directly targeting KIT. Cell Commun Signaling. 2018;16(1):16. doi:10.1186/s12964-018-0228-z
29. Li X, Jiang M, Chen D, et al. miR-148b-3p inhibits gastric cancer metastasis by inhibiting the Dock6/Rac1/Cdc42 axis. J Exp Clin Cancer Res. 2018;37(1):71. doi:10.1186/s13046-018-0729-z
30. He W, Huang L, Li M, Yang Y, Chen Z, Shen X MiR-148b, MiR-152/ALCAM axis regulates the proliferation and invasion of pituitary adenomas cells. Cell Physiol Biochem. 2017;44(2):792–803. doi:10.1159/000485342
31. Agarwal V, Bell GW, Nam JW, Bartel DP Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4.
32. Maskey N, Li D, Xu H, et al. MicroRNA-340 inhibits invasion and metastasis by downregulating ROCK1 in breast cancer cells. Oncol Lett. 2017;14(2):2261–2267 doi:10.3892/ol.2017.6439
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.