Back to Journals » Cancer Management and Research » Volume 12
LncRNA MIR503HG Inhibits Non-Small Cell Lung Cancer Cell Proliferation by Inducing Cell Cycle Arrest Through the Downregulation of Cyclin D1
Authors Xu S, Zhai S, Du T, Li Z
Received 15 August 2019
Accepted for publication 31 January 2020
Published 5 March 2020 Volume 2020:12 Pages 1641—1647
DOI https://doi.org/10.2147/CMAR.S227348
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Shufen Xu,* Shengping Zhai,* Tiantian Du,* Zhan Li
Respiratory Department, Yantai Yuhuangding Hospital Affiliated to Qingdao University, Yantai City, Shandong Province 264000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhan Li
Respiratory Department, Yantai Yuhuangding Hospital Affiliated to Qingdao University, NO. 20 Yuhuangding East Road, Zhifu District, Yantai City, Shandong Province 264000, People’s Republic of China
Email [email protected]
Introduction: LncRNA MIR503HG has been reported to participate in liver cancer and ALK-negative anaplastic large-cell lymphoma, while its role in non-small cell lung cancer (NSCLC) is unknown. We therefore investigated the functions of lncRNA MIR503HG in NSCLC.
Methods: MIR503HG expression in paired cancer and non-cancer tissues from NSCLC patients was analyzed by RT-qPCR. The interaction between cyclin D1 and MIR503HG was analyzed by overexpression experiments. Cell cycle analysis was performed by flow cytometry. Cell proliferation was analyzed by CCK-8 assay.
Results: MIR503HG was downregulated in NSCLC and low levels of MIR503HG were associated with poor survival. In contrast, cyclin D1 was upregulated in NSCLC, and cyclin D1 and MIR503HG were inversely correlated. In NSCLC cells, overexpression experiments revealed that MIR503HG functioned as an upstream inhibitor of cyclin D1. MIR503HG overexpression led to G1 cell cycle arrest, while overexpression of cyclin D1 attenuated the effects of MIR503HG overexpression. Similarly, MIR503HG overexpression resulted in reduced cell proliferation rate, while overexpression of cyclin D1 caused the increased cell proliferation rate and attenuated effects of MIR503HG overexpression.
Conclusion: MIR503HG inhibits NSCLC cell proliferation by inducing cell cycle arrest through the downregulation of cyclin D1.
Keywords: non-small cell lung cancer, lncRNA MIR503HG, cyclin D1, cell cycle, survival
Introduction
The latest cancer statistic data showed that lung cancer is the secondary most common types of malignancy in both men (following prostate cancer) and women (following breast cancer), while lung cancer is the most common cause of cancer-related mortality in both men and women.1 The major cause of the high mortality rate of lung cancer is the low early diagnostic rate.2,3 Therefore, most lung cancer patients are diagnosed at advanced stages, which are not suitable for radical surgery.4 As the most common subtypes of lung cancer, non-small cell lung cancer (NSCLC) accounts for more than 85% of lung cancer cases.5 Up to now, pathogenesis of NSCLC is still largely unclear, which is a big challenge for clinical treatment.6
Accelerated cell cycle progression is the basis of the growth of tumors, and inhibition of cancer cell cycle progression is considered as a promising therapeutic approach for cancer therapy.7 Cyclins, such as cyclin D1, mediates cell phase transitions to promote cancer cell division.8 It also has been shown that cell cycle progression in cancer is also regulated by certain long (>200nt) non-coding RNAs (lncRNAs),9,10 which are not involved in protein-coding but regulate gene expression to participate in diverse cellular processes.11,12 LncRNA MIR503HG plays tumor suppressive roles in liver cancer but promotes ALK-negative anaplastic large-cell lymphoma.13,14 In this study, we investigate the function of MIR503HG in NSCLC.
Materials and Methods
Study Patients
We enrolled 64 NSCLC patients (gender: 39 males and 25 females; age: 35 to 67 years; mean: 52.1±7.1 years) in this study. Those patients were selected from the 144 NSCLC patients admitted by Yantai Yuhuangding Hospital Affiliated to Qingdao University from August 2011 to August 2013. Inclusion criteria: 1) patients confirmed by histopathological biopsy; 2) new cases. Exclusion criteria: 1) patients transferred from other hospitals; 2) any therapies were initiated; 3) recurrent NSCLC; 4) any other diseases were observed. Based on AJCC criteria, there were 14, 15, 18 and 17 cases at stage I-IV, respectively. All patients were informed with the experimental principle. Aforementioned hospital Ethics Committee approved this study.
Follow-Up
A 5-year follow-up was performed after the admission of patients. The follow-up was performed in a monthly manner through outpatient visit and/or phone call. The causes of deaths were recorded and the ones died of other causes or who were lost were not included.
Patients’ Specimens and Cells
NSCLC (cancer) and non-cancer tissues were obtained from each patient during biopsy. Weight of tissues ranged from 0.05 to 0.11g. All tissues were confirmed by at least three pathologists.
Human NSCLC cell lines H1581 and H1993 (ATCC, USA) were used in this study. RPMI-1640 medium (10% FBS) was used to cultivate cells. Cell culture conditions were 37 °C and 5% CO2.
Cell Transfections
MIR503HG and cyclin D1 expression vectors were constructed using pcDNA3 vector by Sangon (Shanghai, China). H1581 and H1993 cells were harvested at confluence of 70–80%. Lipofectamine 2000 (Thermo Fisher Scientific) was used to transfect 10 nM MIR503HG and cyclin D1 expression vector or 10 nM empty pcDNA3 vector (negative control, NC) into 105 cells. Control group included cells without any treatment. Cells were harvested at 24 h post-transfections to perform following experiments.
RT-qPCR
H1581 and H1993 cells (collected at 24 h post-transfection) were mixed with RNAzol reagent (Sigma-Aldrich, USA) with a ratio of 1 mL RNAzol reagent per 105 cells. Tissues were ground in liquid nitrogen and 0.05 g tissue was mixed with 1 mL RNAzol reagent to extract total RNA. All RNA samples were subjected to DNase I digestion. Following reverse transcriptions performed using AMV Reverse Transcriptase XL (Clontech, USA), qPCR mixtures were prepared using the SYBR® Green master mix (Bio-Rad, USA). The expression of MIR503HG and cyclin D1 mRNA was detected using 18S rRNA and GAPDH as endogenous control. All data were processed using 2−ΔΔCT method.
Cell Cycle Analysis
Trypsinization was performed on H1581 and H1993 cells harvested at 24 h after transfections. Pre-cold PBS was used to wash the cells, following by incubation for 3 h with 75% ethanol at 4 °C. After that, cells were washed with fresh pre-cold PBS to remove ethanol. BD Pharmingen™ PI/RNase was then used to incubate with cells in dark for 40 min at 25 °C. Finally, cells were separated by FACSCantoTM Flow Cytometer (BD Biosciences, USA). For each sample, 105 events were counted.
Cell Proliferation Ability Measurement
Eagle’s Minimum Essential Medium (10% FBS) was used to dissolve H1581 and H1993 cells harvested at 24 h after transfections with a ratio of 1 mL cell culture medium per 4×104 cells. Ninety-six-well plates were used to cultivate cells (4×103 cells in 0.1 mL cell suspension per well) under conditions of 37 °C and 5% CO2. CCK-8 solution (10 µL, Sigma,-Aldrich, USA) was added at 4 h before the end of cell culture. Cell culture was stopped at 24, 48, 72 and 96 h. OD values were measured at 450 nm to reflect cell proliferation rates.
Western Blot
RIPA solution (Thermo Fisher Scientific) was used to extract total protein from H1581 and H1993 cells harvested at 24 h after transfections with a ratio of 1 mL c RIPA solution per 2×105 cells. Protein samples were quantified using BCA method and protein samples with good quality were denatured and subjected to 10% SDS-PAGE gel electrophoresis. Protein samples were transferred to PVDF membranes, followed by incubation with 5% non-fat milk (25 min at 25 °C) for blocking. After that, GAPDH (ab9485, 1:1200, Abcam) and cyclin D1 antibody (ab226977, 1:1200, Abcam) rabbit polyclonal primary antibodies were used to incubate with PVDF membranes overnight at 4 °C. The next day, PVDF membranes were further incubated with IgG-HRP secondary antibody (1:800, goat anti-rabbit, MBS435036, MyBioSource) for 2 h at room temperature. Image J v1.46 software was used to process all data.
Statistical Analysis
Data in this paper were all mean values. Mean values were from data of at least three biological replicates of each experiment. Differences between two types of tissues were analyzed using paired t-test. Differences among multiple groups were analyzed by performing ANOVA (one-way) and Tukey’s test. Linear regression was used for correlation analysis. The 64 NSCLC patients were grouped into high and low expression group based on the median expression level of MIR503HG in NSCLC tissues (n=32). The cutoff value was 1.97. K-M method was used to plot survival curves, and Log rank test was used to compare survival curves. Differences were significant when p<0.05.
Results
MIR503HG and Cyclin D1 Were Dysregulated in NSCLC
We first analyzed the differential expression of MIR503HG and cyclin D1 in NSCLC by performing RT-qPCR followed by paired t-test. The data showed that, comparing to non-cancer tissues, MIR503HG was significantly downregulated (Figure 1A), and cyclin D1 was significantly upregulated (Figure 1B) in NSCLC tissues collected from the 64 NSCLC patients (p<0.05).
MIR503HG Expression in NSCLC Predicted Survival
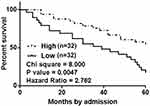
Based on the methods aforementioned, survival curves were plotted for both low and high MIR503HG level groups and compared between these two groups. It was observed that patients in low MIR503HG level group had significantly lower overall survival rate comparing to patients in high MIR503HG level group (Figure 2).
Cyclin D1 and MIR503HG Were Inversely Correlated in NSCLC Tissues
Linear regression was performed to analyze the correlations between Cyclin D1 and MIR503HG. It was observed that cyclin D1 and MIR503HG were significantly correlated in NSCLC tissues (Figure 3A). However, the correlation between Cyclin D1 and MIR503HG in non-cancer tissues was not significant (Figure 3B).
MIR503HG Was an Upstream Inhibitor of Cyclin D1 in NSCLC Tissues
MIR503HG and cyclin D1 were transfected into H1581 and H1993 cells and expression levels of MIR503HG and cyclin D1 were significantly increased at 24 h after transfections comparing to control (C) and negative control (NC) groups (Figure 4A, p<0.05). Moreover, cells with MIR503HG overexpression showed significantly downregulated cyclin D1 compared to two controls (Figure 4B, p<0.05), while cells with cyclin D1 overexpression showed no significantly altered expression of MIR503HG (Figure 4C).
MIR503HG Inhibits NSCLC Cell Proliferation Through Cyclin D1
Comparing to NC and C two controls, MIR503HG overexpression led to increased percentage of cells at G1 phase and decreased percentage of cells at G2 phase. In addition, cyclin D1 overexpression played an opposite role and attenuated the effects of MIR503HG overexpression (Figure 5A, p<0.05). Comparing to the two controls, MIR503HG overexpression resulted in reduced cell proliferation rate, while overexpression of cyclin D1 caused the increased cell proliferation rate and attenuated effects of MIR503HG overexpression (Figure 5B, p<0.05).
Discussion
We in this paper discussed the function and clinical potentials of MIR503HG for NSCLC. We found that MIR503HG was downregulated in NSCLC and inhibited NSCLC cell cycle progression by downregulating cyclin D1. We also found that MIR503HG may be used to assist the prognosis of NSCLC.
The same lncRNA may have opposite functions in different types of cancer. For instance, lncRNA TUG1 promote cancer cell prognosis in glioma, indicating its tumor suppressive function.15 However, TUG1 in osteosarcoma promote cancer cell proliferation, indicating its oncogenic role.16 MIR503HG was downregulated in liver cancer and inhibits tumor metastasis by inactivating oncogenic NF-κB pathway.13 However, in another study, Huang et al showed that MIR503HG can enhance the proliferation of cancer cell in ALK-negative anaplastic large-cell lymphoma.14 In this study, we observed the downregulated MIR503HG in NSCLC and the accelerated cancer cell proliferation after MIR503HG overexpression. Our data suggest the tumor suppressive role of MIR503HG in NSCLC.
In liver cancer, the downregulated MIR503HG indicates the poor survival of cancer patients.13 Accurate prognosis is also critical for NSCLC patients due to the fact that most NSCLC patients are diagnosed at late stages.2,3 Therefore, accurate prognosis may guide the individualized treatment program, thereby prolonging patients’ survival. We showed that low MIR503HG level in NSCLC tissues was closely associated with the poor survival of NSCLC patients. Therefore, detecting the expression of MIR503HG in NSCLC tissue may assist the prognosis of NSCLC.
Cyclin D1 in cancer development and progression can be regulated by certain lncRNAs.17,18 We in this study found that MIR503HG was likely an upstream inhibitor of cyclin D1, and the regulation of cyclin D1 by MIR503HG is involved in the regulation of NSCLC cell cycle progression and cell proliferation. Therefore, overexpression of MIR503HG may be a potential target to inhibit NSCLC tumor growth. However, clinical studies are needed to verify our hypothesis.
In conclusion, MIR503HG was downregulated in NSCLC and overexpression of MIR503HG may suppress cancer progression by inhibiting cell cycle progression through the downregulation of cyclin D1.
Ethics
All patients provided written informed consent and this study was conducted in accordance with the Declaration of Helsinki.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.v69.1
2. Canadian Task Force on Preventive Health Care. Recommendations on screening for lung cancer. CMAJ. 2016;188(6):425–432. doi:10.1503/cmaj.151421
3. Gasparri R, Santonico M, Valentini C, et al. Volatile signature for the early diagnosis of lung cancer. J Breath Res. 2016;10(1):016007. doi:10.1088/1752-7155/10/1/016007
4. Sperduto PW, Yang TJ, Beal K, et al. Estimating survival in patients with lung cancer and brain metastases: an update of the graded prognostic assessment for lung cancer using molecular markers (Lung-molGPA). JAMA Oncol. 2017;3(6):827–831. doi:10.1001/jamaoncol.2016.3834
5. Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet. 2011;378(9804):1727–1740. doi:10.1016/S0140-6736(10)62101-0
6. Aisner DL, Marshall CB. Molecular pathology of non–small cell lung cancer: a practical guide. Am J Clin Pathol. 2012;138(3):332–346. doi:10.1309/AJCPFR12WJKCEEZZ
7. Rehman J, Zhang HJ, Toth PT, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26(5):2175–2186. doi:10.1096/fj.11-196543
8. Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–3093. doi:10.1242/dev.091744
9. Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3):e1003368. doi:10.1371/journal.pgen.1003368
10. Zhang K, Sun X, Zhou X, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6(1):537–546. doi:10.18632/oncotarget.2681
11. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606
12. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi:10.1038/nrm3679
13. Wang H, Liang L, Dong Q, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018;8(10):2814–2829. doi:10.7150/thno.23012
14. Huang P-S, Chung I, Lin Y-H, et al. The long non-coding RNA MIR503HG enhances proliferation of human ALK-negative anaplastic large-cell lymphoma. Int J Mol Sci. 2018;19(5):1463. doi:10.3390/ijms19051463
15. Li J, Zhang M, An G, et al. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp Biol Med (Maywood). 2016;241(6):644–649. doi:10.1177/1535370215622708
16. Yun-Bo F, Xiao-Po L, Xiao-Li L, et al. LncRNA TUG1 is upregulated and promotes cell proliferation in osteosarcoma. Open Med (Wars). 2016;11(1):163–167. doi:10.1515/med-2016-0031
17. Liu Y, Wang -L-L, Chen S, et al. LncRNA ABHD11‐AS1 promotes the development of endometrial carcinoma by targeting cyclin D1. J Cell Mol Med. 2018;22(8):3955–3964. doi:10.1111/jcmm.2018.22.issue-8
18. Zhu L, Di Y, Liu G, et al. LncRNA CRNDE promotes hepatocellular carcinoma cell proliferation and upregulates cyclin D1 expression. Int J Clin Exp Med. 2018;11(7):6957–6964.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.