Back to Journals » Cancer Management and Research » Volume 12
LncRNA MALAT1 Promotes the Proliferation, Migration, and Invasion of Melanoma Cells by Downregulating miR-23a
Authors Wang P, Hu L, Fu G, Lu J, Zheng Y, Li Y, Jia L
Received 11 February 2020
Accepted for publication 17 June 2020
Published 29 July 2020 Volume 2020:12 Pages 6553—6562
DOI https://doi.org/10.2147/CMAR.S249348
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Pan Wang,1,* Liu Hu,2,* Guili Fu,1 Jingjing Lu,1 Yuanquan Zheng,1 Ying Li,2 Lin Jia3
1Department of Dermatology, Wuhan Children’s Hospital (Wuhan Maternal and Child Healthcare Hospital), Tongji Medical College, Huazhong University of Science & Technology, Wuhan 430015, Hubei Province, People’s Republic of China; 2Department of Radiotherapy, Hubei Cancer Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei Province, 430071, People’s Republic of China; 3Department of Nephrology, The Central Hosptial of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430015, Hubei Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ying Li
Department of Radiation Oncology, Hubei Cancer Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430071, Hubei Province, People’s Republic of China
Tel +86 13628635363
Email [email protected]
Lin Jia
Department of Nephrology, The Central Hosptial of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430015, Hubei Province, People’s Republic of China
Tel +86 13476126250
Email [email protected]
Purpose: This study was designed to investigate the relationship between long-chain non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (lncRNA MALAT1)/miR-23a-23a and melanoma.
Patients and Methods: Fifty-two cases of corresponding non-tumor normal tissues and 109 cases (including 62 cases of primary melanoma and 47 cases of metastatic melanoma) were collected. Real-time fluorescent PCR quantified lncRNA MALAT1 and miR-23a, and counted the 3-year survival of high/low miR-23 and high/low lncRNA MALAT1 populations. We predicted the binding site according to the sequence information of lncRNA MALAT1 and miR-23a. lncRNA MALAT1 siRNA and miR-23a mimics vectors were constructed and transfected into melanoma cell lines respectively to observe their effects on cells.
Results: Compared with corresponding non-tumor normal tissues, lncRNA MALAT1 in melanoma tissue increased while miR-23a decreased. Compared with primary melanoma, metastatic melanoma was higher and miR-23a was lower. Downregulation of lncRNA MALAT1 caused upregulation of miR-23a, and lncRNA MALAT1 could bind to miR-23a. Downregulating lncRNA MALAT1 or upregulating miR-23a inhibited cell proliferation, migration and invasion and promoted apoptosis. Rescue experiments revealed that downregulation of miR-23a could offset cell changes caused by downregulation of lncRNA MALAT1.
Conclusion: lncRNA MALAT1 promotes malignant proliferation of melanoma cells through miR-23a.
Keywords: melanoma, long-chain non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (lncRNA MALAT1), miR-23a, malignant proliferation
Introduction
Melanoma is a malignant tumor type caused by malignant transformation of melanocytes, which can be divided into cutaneous melanoma and extracutaneous melanoma according to histological types.1 In the past 50 years, the morbidity and mortality of melanoma have increased year by year.2 In 2018, its morbidity and mortality in Australia were the highest in the world respectively.3 High recurrence rate, high drug resistance and strong metastasis are its main characteristics.1,4 Ultraviolet, race, human immunodeficiency virus, gene, and telomere length are important risk factors for melanoma.5–9 One of the biggest difficulties in clinical treatment is how to evaluate and predict cancer metastasis and death.10 Specific biomarkers with high sensitivity are the key to solve this problem. Non-coding RNA is an important link in cancer gene regulation, so it may have potential marker value. Manly studies have revealed that non-coding RNAs such as miR-10b, miR-1246, and miR-206 can be used as biomarkers of melanoma.11–13 Studying the relationship between non-coding RNA and cancer is helpful for the discovery of melanoma biomarkers.
Long-chain non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (lncRNA MALAT1) is a vital participant in most cancers. 3ʹ non-coding region of lncRNA MALAT1 sequence has multiple sequence sites that can be combined with different target genes, and through these sites, target gene expression can be regulated, cell phenotype can be changed, and tumor formation and development can be caused or inhibited. Many studies14–19 have shown that lncRNA MALAT1 is relevant to the occurrence and development of cancer. miR-23a is a 73bp miRNA located on human chromosome 19. Mechanism of action of miR-23a is similar to that of lncRNA MALAT1, which also obstructs its post-transcriptional process by binding to specific sequences of downstream target genes. miR-23a is an active tumor regulatory element, which can regulate the formation and development of most malignant tumors through different genes.20–24
Previous studies25–28 manifest that increased lncRNA MALAT1 or decreased miR-23a promote melanoma tumor formation and metastasis. lncRNA MALAT1 is upregulated and miR-23a is downregulated in the detection of melanoma tissue samples, while the Starbase2.0 database predicts that the two have specific pairing binding sites. Based on this, it is speculated that they may participate in the occurrence and development of melanoma through binding and pairing. At present, there is no research showing that lncRNA MALAT1 and miR-23a can participate in melanoma, so this article will study how the two jointly regulate melanoma.
Materials and Methods
Melanoma Patients and Tissue Samples
From February 2012 to April 2014, 109 cases (including 62 cases of primary melanoma and 47 cases of metastatic melanoma) of melanoma tissues and 52 cases of corresponding non-tumor normal tissues were collected. Inclusion criteria for melanoma patients were as follows: those diagnosed as melanoma according to clinical features or pathological sections. Exclusion criteria were as follows: patients with other tumors; those who had a previous history of radiotherapy, chemotherapy or surgical resection; those combined with other skin diseases. The hospital informed the patients of the research information throughout the study, and this was approved by the ethics Committee of the Central Hospital of Wuhan, which is in line with the Declaration of Helsinki. Tissue samples and sections were stored at −80°C for testing. In this study, the discharged patients were followed up for 3 years by telephone or on-site visit. Definition of survival time was from diagnosis of melanoma to death. All patients in our study provided informed consent. All participates provided their written informed consent prior to study inclusion.
Cell Culture and Transfection
Melanoma cell lines A375, A2058, SK-MEL-5, SK-MEL-28 and human normal melanoma cell lines (MC) were purchased from the cell bank of the American Committee for Type Culture Collection. The cell culture medium consists of 10% fetal bovine serum solution (Gibco), 1% penicillin/streptomycin solution (100x, Solarbio) and DMEM basal medium (Hyclone). Cells were cultured in animal cell incubator (Binder, Germany) at 37°C and 5%CO2 until they were in a good growth condition. Before transfection, the medium was replaced with fetal bovine serum-free medium. During transfection, 1×105 cells per well were inoculated into 6-well plates. The miR-23a mimics, miR-23a inhibitor, MALAT1 siRNA and corresponding negative control vectors were all designed and synthesized by Shanghai Sangon Biotech Co., Ltd. Cell lines were transfected with Lipofectamine 2000 transfection kit (Invitrogen, USA). The procedures referred to the kit instructions. Eight hours after transfection, fresh culture medium was changed to avoid poisoning cells.
qPCR
Trizol method was employed to extract total RNA from tissues or cells. OD value of total RNA was obtained at 260–280 nm, and OD260/OD280>1.8 was used for subsequent RT-PCR quantification. FastKing One-Step Reverse Transcription-Fluorescence Quantitative Kit (Tiangen, Beijing, China) performed reverse transcription and amplification on total RNA samples. miR-23a and lncRNA MALAT1 primers were designed and synthesized by Tiangen, Beijing. The qPCR reaction system (50 μL) was as follows: upstream primer 1.25 μL, downstream primer 1.25 μL, probe 1.0 μL, RNA template 10 pg/μg, 50×ROX Reference Dye ROX 5 μL, and RNase-Free ddH2O added to the total reaction system of 50 uL. Reaction process was as follows: reverse transcription at 50°C for 30 min and circulation once; pre-denaturing at 95°C for 3 min and circulating once; denaturing at 95°C for 15 s, annealing at 60°C for 30s, and circulating 40 times. Results were analyzed by ABI PRISM 7000 instrument (Applied Biosystems, USA). The internal reference genes were U6 and GAPDH, which were standardized by 2–ΔΔCt method. miR-23a: forward 5ʹ- CGC GAT CAC ATT GCC AGG G −3ʹ, reserve 5ʹ- GTG CAG GGT CCG AGG T −3ʹ, MALAT1: forward 5ʹ- GAC GGA GGT TGA GAT GAA GC −3ʹ, reserve 5ʹ-ATT CGG GGC TCT GTA GTC CT-3ʹ.
Western Blot
Melanoma cells were washed twice with ice-cold PBS buffer, and RIPA lysis buffer (containing 150 mmol/L NaCl, 0.1% Triton X-100, 50 mmol/L Tris-HCl with pH of 8.0 and protein inhibitor) was added to them, all of which were purchased from Solarbio. We repeatedly blew the buffer until they were completely lysed. The solution was centrifuged at 1.6×104×g in a precooled centrifuge at 4°C for 20 min, and the precipitate was discarded. We took 50 μL supernatant and used BCA kit (Thermo Fisher Scientific) to determine the protein concentration. The protein was separated by SDS-PAGE electrophoresis with a sample loading of 20–30 μg. It was transferred to polyvinylidene fluoride membrane (EMD millipore) and 5% skim milk-PBS buffer blocked the protein for 1 h at room temperature. Subsequently, the protein to be detected and β-actin primary antibody were supplemented and left to stand all night long at 4°C. The PBS buffer was used to clean polyvinylidene fluoride membrane for three times, then goat anti-rabbit secondary antibody (HRP cross-linked) was supplemented, and left to stand at room temperature for 1 h. Finally, PBS solution was employed to wash polyvinylidene fluoride membrane and ECL luminescent solution was used for visualization. The internal reference protein was β-actin, and the relative expression level of the protein to be detected = gray value of the band to be detected/gray value of the β-actin band. Protein primary antibody and goat anti-rabbit secondary antibody were purchased from Abcam.
Transwell Method
Cells were inoculated into the migration upper chamber with 2×104 cells/well (200 μL of mixed solution containing 10% fetal bovine serum and 1%DMEM medium were added to the upper chamber in advance), and DMEM medium (containing 10% fetal bovine serum, with a total volume of 500 μL) was supplemented to the lower chamber. The transfer cell was cultured in 37°C and 5%CO2 for 24 h, then the upper chamber liquid was removed and the cell wall was wiped off. Four per cent paraformaldehyde immobilized cells of the reverse side in Transwell chamber for 20 min. They were stained by crystal violet for 15 min, and Transwell chamber was cleaned by PBS buffer solution. Photographs of cell migration were collected under a 200-fold microscope. The cell number was calculated by randomly selected 3 fields of view, and the average value was taken as the number of transmembrane cells. The test was repeated three times. Invasion was paved with 8% matrix glue on the above steps, and the number of cells per well was increased to 5×104.
MTT Assay
We took 96-well plates, inoculated 3×103 transfected cell lines per well, inoculated 4 well plates in total, and inoculated 3 wells per well plate. Cells were cultured at 37°C/5%CO2. One well plate was taken out every 24 h, 10 μL of 5 mg/mL MTT solution was added to each well, the culture medium was removed after the culture was continued for 4 h at 37°C/5%CO2, 100 μL of dimethyl sulfoxide (Solarbio) was supplemented, and the OD value of the solution at 570 nm was measured with an enzyme reader after shaking.
Dual Luciferase Reporter Gene
LncRNA MALAT1 and miR-23a were sequence matched using Starbase3.0 to predict their binding sites. Cells were inoculated into 12-well plates and cultured until they grew well. PmirGLO-MALAT1-wt (including binding sites) and pmirGLO-MALAT1-mut (binding site mutation) were constructed according to the predicted sites and pmirGLO was set as the control group. The above three were co-transfected with NC mimics and miR-137 mimics respectively. After 48 h, the culture medium was removed, the PBS buffer was used to wash the cells, and the luciferase intensity was detected by a dual luciferase reporter gene assay system (Promega). Relative luciferase intensity = firefly luciferase intensity/sea pansy luciferase intensity.
Statistics and Analysis
The experiment was repeated 3 times, and the measurement results were expressed as Mean±SD (standard deviation). SPSS 20.0 software (IBM Company, USA) was used for data difference analysis and Graphpad 8.0 was used for data picture drawing. The differences between the two groups were compared by independent-samples T-test. Those among groups were analyzed via one-way analysis of variance, and post hoc pairwise comparison was under LSD-t-test. Log-rank (Mantel-Cox) test was used to compare the differences between survival curves. All comparisons were two-tailed tests. Taking 95% as its confidence interval, the differences were statistically significant when p was lower than 0.05.
Results
LncRNA MALALT1 and miR-23a are Associated with Melanoma
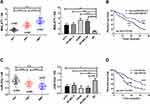
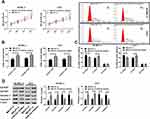
In this article, we collected 52 corresponding non-tumor normal tissues and 109 melanoma tissues (including 62 primary melanoma and 47 metastatic melanoma). qPCR quantified lncRNA MALAT1 and miR-23a levels. Figures 1A and C indicate that lncRNA MALAT1 and miR-23a of metastatic melanoma are the highest and lowest respectively in the three groups. Their expression levels in cells were also detected, and the results were similar to those of tissues. Figures 1B and D indicate that elevated lncRNA MALAT1 or reduced miR-23a is associated with a 3-year low survival rate for melanoma patients. The above results reveal that lncRNA MALAT1 and miR-23a increases and decreases in melanoma respectively, and are closely related to the survival of melanoma patients.
LncRNA MALAT1 Promotes Malignant Proliferation, Migration and Invasion of Melanoma Cells
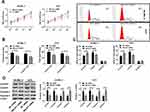
SK-MEL-5 cells and A375 cells were selected as research objects in this article. lncRNA MALAT1 was downregulated to study the relationship between lncRNA MALAT1 and melanoma. Cell proliferation was detected by MTT method, cell cycle was detected by flow cytometry, protein level was tested via Western blot method, and cell migration and invasion were checked by the Transwell method. Figures 2A and B indicate that downregulation of lncRNA MALAT1 inhibits cell proliferation, invasion and migration. When cell proliferation was inhibited, S-phase cells decreased while G1-phase cells increased, accompanied by downregulation of CyclinD1 and CyclinE1. When apoptosis increased, apoptosis proteins Caspase 3 and Caspase 9 were upregulated, so the two could be used to evaluate apoptosis. Figure 2C and D indicate that downregulation of lncRNA MALAT1 causes G1 phase cells to increase and S phase cells to decrease by inhibiting CyclinD1 and CyclinE1. Downregulation of lncRNA MALAT1 promotes apoptosis by upregulating Caspase 3 and Caspase 9.
miR-23a Inhibits Malignant Proliferation, Migration and Invasion of Melanoma Cells
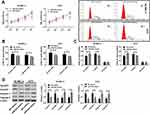
Results 2.1 shows that miR-23a was correlated to melanoma. In this article, the effect of miR-23a on melanoma was studied by upregulating it. Figure 3 reveals that upregulated miR-23a inhibits cell proliferation by downregulating CyclinD1\E1 to increase G1-phase cells and decrease S-phase cells. Upregulated miR-23a promotes apoptosis by upregulating Caspase 3 and Caspase 9. Upregulated miR-23a inhibits cell migration and invasion. The above results manifest that miR-23a inhibits malignant proliferation, migration and invasion of melanoma cells, and its upregulation can inhibit the survival of melanoma cells.
miR-23a is the Target Gene of lncRNA MALAT1
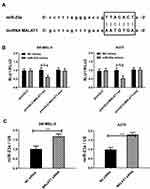
Figures 2 and 3 display that downregulation of lncRNA MALAT1 and up-regulation of miR-23a can have similar effects on melanoma cells, while downregulation of lncRNA MALAT1 causes downregulation of miR-23a (Figure 4C). Hence, we speculate that miR-23a may be the target gene of lncRNA MALAT1. To verify this conjecture, the sequence matching and analysis of the two were carried out by using starbase2.0. Figure 4A reveals that the binding site of miR-23a exists in the 3ʹ non-coding region of lncRNA MALAT1. According to this prediction site, pmirGLO-MALAT1-wt and pmirGLO-MALAT1-mut were constructed and co-transfected into melanoma cells in NC mimics and miR-23amamimcs respectively. Figure 4B shows that when miR-23ama mimics is co-transfected with pmirGLO-MALAT1-wt, the relative activity of luciferase decreases statistically, which indicates that lncRNA MALAT1 can target to inhibit miR-23a. The above results indicate that miR-23a is the downstream target gene of lncRNA MALAT1.
Rescue Experiment
MALAT1 siRNA and MALAT1 siRNA+miR-23a inhibitor were transfected into melanoma cancer cells respectively to observe cell changes. Figure 5 shows that downregulation of miR-23a counteracts cell proliferation, migration and invasion reduction caused by downregulation of MALAT1, Caspase 3 and Caspase 9 are upregulated, CyclinD1 and CyclinE1 are downregulated, S-phase cells are reduced and G1-phase cells are increased. The above results suggest that lncRNA MALAT1 promotes malignant proliferation, migration and invasion of cells and inhibits apoptosis by downregulating miR-23a.
Discussion
Pathology and risk stratification are important factors affecting the prognosis of melanoma patients.1 lncRNA or miRNA may be helpful to assess the risk stratification of melanoma. Changes in cellular biological functions do not depend solely on genetic information stored on gene fragments. Epigenetic means involving non-coding RNA are also momentous links in changing biological functions. In the formation and development of melanoma, melanocytes not only gain immortalization and infinite proliferation, but can also be transferred to other places to invade normal cells. In this process, melanocyte genes are jointly regulated by lncRNA and miRNA, so studying the relationship between the two is helpful to obtain important information about melanoma development.4
When comparing primary melanoma tissues, metastatic melanoma tissues and corresponding non-tumor normal tissues, we found that melanoma tissues showed high lncRNA MALAT1 and low miR-23a compared with corresponding non-tumor normal tissues. As melanoma is a malignant tumor with strong metastasis,1 melanoma tissue is subdivided into primary melanoma and metastatic melanoma in this article. The results reveal that lncRNA MALAT1 and miR-23a are expressed higher and lower in metastatic melanoma respectively. In addition, lncRNA MALAT1 and miR-23a also showed similar trends in cells. Li et al26 and Ma et al28 have found that lncRNA MALAT1 was upregulated and miR-23a was downregulated in melanoma, respectively. According to the results of this study and other studies, it can be inferred that increased lncRNA MALAT1 and decreased miR-23a may be related to melanoma. The 3-year follow-up results (Figures 1B and D) indicate that the survival conditions of the high lncRNA MALAT1 group and the low miR-23a group are worse than those of the low lncRNA MALAT1 group and the high miR-23a group. These results suggest that lncRNA MALAT1 and miR-23a may be involved in melanoma.
The relationship between lncRNA MALAT1, miR-23a and melanoma was studied by regulating them. Cell proliferation was detected by MTT method, cell cycle was detected by flow cytometry, cell migration and invasion were detected by Transwell method, and CyclinD1 and CyclinE1 and apoptosis protein Caspase 3 and Caspase 9 were tested by Western blot method. The results revealed that downregulation of lncRNA MALAT1 or up-regulation of miR-23a not only downregulated CyclinD1 and CyclinE1 and up-regulated Caspase 3 and Caspase 9, but also resulted in cell proliferation and S-phase cell decrease, G1-phase cell increase, and cell migration and invasion ability decrease. CyclinD1 and CyclinE1 are proteins that monitor the transition from G1 phase to S phase.29,30 If the expression or activity of both proteins changes, cell cycle will be disturbed. In this article, the downregulation of CyclinD1 and CyclinE1 causes the increase of G1-phase cells and the decrease of S-phase cells. For the cell population with vigorous proliferation, S-phase cells should occupy the majority. Downregulation of CyclinD1 and Cyclin 1 can block cells in the G1 phase, bringing about a decrease in cell proliferation. The MTT method also shows that the downregulation of CyclinD1 and Cyclin 1 is accompanied by a decrease in cell proliferation. Apoptosis proteins Caspase 3 and Caspase 9 can be employed to evaluate apoptosis. Upregulation of Caspase 3 and Caspase 9 caused by downregulation of lncRNA MALAT1 or upregulation of miR-23a indicates that apoptosis increases obviously. According to the above results, it is speculated that in the occurrence and development of melanoma, elevated lncRNA MALAT1 and lowered miR-23a promote malignant cell proliferation and inhibit apoptosis by upregulating CyclinD1\E1 and downregulating Caspase 3\9.
miR-23a increases when lncRNA MALAT1 is downregulated, which may suggest that the two may jointly affect melanoma cells through some action. Many studies have shown that lncRNA can regulate its expression level by binding with miRNA.17–20 Therefore, this article analyzes and studies whether there is a binding site between lncRNA MALAT1 and miR-23a. Targetscan7.2 analysis results reveal that the 3ʹ non-coding region of lncRNA MALAT1 has a site matching and binding with miR-23a, while the dual luciferase reporter gene indicates that lncRNA MALAT1 targets and inhibits miR-23a in melanoma cells. The above results indicate that the decrease of miR-23a in melanoma is due to elevated lncRNA MALAT1. According to Results 2.2–2.5, lncRNA MALAT1 promotes malignant proliferation, migration and invasion of melanoma cells by downregulating miR-23a.
After studying the effects of lncRNA MALAT1 and miR-23a on cell biological functions, it is found that their combination causes melanoma cells to acquire malignant proliferation ability and inhibit apoptosis. lncRNA MALAT1 participates in melanoma by regulating miR-23a, and what mechanism does miR-23a regulate melanoma? Since many studies27–29 suggest that miR-23a participates in disease occurrence by regulating target genes, which genes are downstream of lncRNA MALAT1/miR-23a regulatory axis? To solve this problem, we will further explore the relationship between miR-23a and a target gene downstream in future research. Besides, in view of the influence of lncRNA MALAT1 and miR-23a on melanoma cells, this article speculates that the two may be helpful to assess the risk stratification and prognosis of melanoma, and this will be discussed in subsequent studies.
Conclusion
To summarize, lncRNA MALAT1 is found to promote malignant proliferation of melanoma cells and inhibit apoptosis by downregulating miR-23a. This regulatory mechanism greatly promotes the formation and development of melanoma. Downregulation of lncRNA MALAT1 or up-regulation of miR-23a may be helpful to inhibit its further growth and development.
Funding
This study was financially supported by 2018 Wuhan Medical Research Project (WX18Q45).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chopra A, Sharma R, Rao UNM. Pathology of melanoma. Surg Clin North Am. 2020;100(1):43–59. doi:10.1016/j.suc.2019.09.004
2. Berwick M, Buller DB, Cust A, et al. Melanoma epidemiology and prevention. Cancer Treat Res. 2016;167:17–49.
3. Carr S, Smith C, Wernberg J. Epidemiology and risk factors of melanoma. Surg Clin North Am. 2020;100(1):1–12. doi:10.1016/j.suc.2019.09.005
4. Arnold J, Engelmann JC, Schneider N, Bosserhoff AK, Kuphal S. miR-488-5p and its role in melanoma. Exp Mol Pathol. 2019;112:104348. doi:10.1016/j.yexmp.2019.104348
5. Facciola A, VenanziRullo E, Ceccarelli M, et al. Malignant melanoma in HIV: epidemiology, pathogenesis, and management. Dermatol Ther. 2019;e13180.
6. Ribero S, Glass D, Bataille V. Genetic epidemiology of melanoma. Eur J Dermatol. 2016;26(4):335–339. doi:10.1684/ejd.2016.2787
7. Dawes SM, Tsai S, Gittleman H, Barnholtz-Sloan JS, Bordeaux JS. Racial disparities in melanoma survival. J Am Acad Dermatol. 2016;75(5):983–991. doi:10.1016/j.jaad.2016.06.006
8. Ghiasvand R, Weiderpass E, Green AC, Lund E, Veierod MB. Sunscreen use and subsequent melanoma risk: a population-based cohort study. J Clin Oncol. 2016;34(33):3976–3983. doi:10.1200/JCO.2016.67.5934
9. Rachakonda S, Kong H, Srinivas N, et al. Telomere length, telomerase reverse transcriptase promoter mutations, and melanoma risk. Genes Chromosomes Cancer. 2018;57(11):564–572. doi:10.1002/gcc.22669
10. Scolyer RA, Rawson RV, Gershenwald JE, Ferguson PM, Prieto VG. Melanoma pathology reporting and staging. Mod Pathol. 2020;33(Suppl 1):15–24. doi:10.1038/s41379-019-0402-x
11. Saldanha G, Elshaw S, Sachs P, et al. microRNA-10b is a prognostic biomarker for melanoma. Mod Pathol. 2016;29(2):112–121. doi:10.1038/modpathol.2015.149
12. Armand-Labit V, Meyer N, Casanova A, et al. Identification of a circulating microRNA profile as a biomarker of metastatic cutaneous melanoma. Acta Derm Venereol. 2016;96(1):29–34.
13. Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. Int J Clin Exp Pathol. 2015;8(3):3097–3103.
14. Arun G, Spector DL. MALAT1 long non-coding RNA and breast cancer. RNA Biol. 2019;16(6):860–863. doi:10.1080/15476286.2019.1592072
15. Gordon MA, Babbs B, Cochrane DR, Bitler BG, Richer JK. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog. 2019;58(2):196–205. doi:10.1002/mc.22919
16. Malakar P, Stein I, Saragovi A, et al. Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR-mediated translation of TCF7L2. Cancer Res. 2019;79(10):2480–2493. doi:10.1158/0008-5472.CAN-18-1432
17. Yu W, Ding J, He M, et al. Estrogen receptor beta promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene. 2019;38(8):1225–1238. doi:10.1038/s41388-018-0463-1
18. Xi Z, Si J, Nan J. LncRNA MALAT1 potentiates autophagy-associated cisplatin resistance by regulating the microRNA30b/autophagy-related gene 5 axis in gastric cancer. Int J Oncol. 2019;54(1):239–248. doi:10.3892/ijo.2018.4609
19. Xie JJ, Li WH, Li X, Ye W, Shao CF. LncRNA MALAT1 promotes colorectal cancer development by sponging miR-363-3p to regulate EZH2 expression. J Biol Regul Homeost Agents. 2019;33(2):331–343.
20. Zhang X, Wu N, Wang J, Li Z. LncRNA MEG3 inhibits cell proliferation and induces apoptosis in laryngeal cancer via miR-23a/APAF-1 axis. J Cell Mol Med. 2019;23(10):6708–6719. doi:10.1111/jcmm.14549
21. Karimi N, Ali HosseinpourFeizi M, Safaralizadeh R, et al. Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J Chin Med Assoc. 2019;82(3):215–220. doi:10.1097/JCMA.0000000000000031
22. Sruthi TV, Edatt L, Raji GR, et al. Horizontal transfer of miR-23a from hypoxic tumor cell colonies can induce angiogenesis. J Cell Physiol. 2018;233(4):3498–3514. doi:10.1002/jcp.26202
23. Yachi K, Tsuda M, Kohsaka S, et al. miR-23a promotes invasion of glioblastoma via HOXD10-regulated glial-mesenchymal transition. Signal Transduct Target Ther. 2018;3(1):33. doi:10.1038/s41392-018-0033-6
24. Chien MH, Lee WJ, Yang YC, et al. KSRP suppresses cell invasion and metastasis through miR-23a-mediated EGR3 mRNA degradation in non-small cell lung cancer. Biochim Biophys Acta Gene Regul Mech. 2017;1860(10):1013–1024. doi:10.1016/j.bbagrm.2017.08.005
25. Sun Y, Cheng H, Wang G, et al. Deregulation of miR-183 promotes melanoma development via lncRNA MALAT1 regulation and ITGB1 signal activation. Oncotarget. 2017;8(2):3509–3518. doi:10.18632/oncotarget.13862
26. Li F, Li X, Qiao L, Liu W, Xu C, Wang X. MALAT1 regulates miR-34a expression in melanoma cells. Cell Death Dis. 2019;10(6):389. doi:10.1038/s41419-019-1620-3
27. Ding F, Lai J, Gao Y, et al. NEAT1/miR-23a-3p/KLF3: a novel regulatory axis in melanoma cancer progression. Cancer Cell Int. 2019;19(1):217. doi:10.1186/s12935-019-0927-6
28. Ma M, Dai J, Tang H, et al. MicroRNA-23a-3p inhibits mucosal melanoma growth and progression through targeting adenylate cyclase 1 and attenuating cAMP and MAPK pathways. Theranostics. 2019;9(4):945–960. doi:10.7150/thno.30516
29. Azevedo-Barbosa H, Ferreira-Silva GA, Silva CF, et al. Phenylpropanoid-based sulfonamide promotes cyclin D1 and cyclin E down-regulation and induces cell cycle arrest at G1/S transition in estrogen positive MCF-7 cell line. Toxicol in vitro. 2019;59:150–160. doi:10.1016/j.tiv.2019.04.023
30. Tang Z, Fang Y, Du R. MicroRNA-107 induces cell cycle arrests by directly targeting cyclin E1 in ovarian cancer. Biochem Biophys Res Commun. 2019;512(2):331–337. doi:10.1016/j.bbrc.2019.03.009
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.