Back to Journals » OncoTargets and Therapy » Volume 13
lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p
Authors Zhang Z, Li M, Zhang Z
Received 30 November 2018
Accepted for publication 30 January 2019
Published 13 February 2020 Volume 2020:13 Pages 1343—1354
DOI https://doi.org/10.2147/OTT.S196619
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Zhenming Zhang,1,* Ming Li,1,* Zhitao Zhang2
1Department II of General Surgery, Zhangye People’s Hospital Affiliated to Hexi University, Zhangye, Gansu, China; 2Department of Oncology, Zhangye People’s Hospital Affiliated to Hexi University, Zhangye, Gansu, China
*These authors contributed equally to this work
Correspondence: Zhitao Zhang
Department of Oncology, Zhangye People’s Hospital Affiliated to Hexi University, No 67, West Huancheng Road, Zhangye, Gansu 734000, China
Tel +86 936 855 3195
Email [email protected]
Objective: Various regulatory mechanisms have been demonstrated to be associated with cancer progression. ncRNA and mRNA play important roles in gastric cancer (GC) cell growth and drug resistance, respectively. However, the regulatory network of ncRNA and mRNA in GC oxaliplatin (OXA) resistance has not been fully clarified.
Methods: The expression of miR-22-3p, MALAT1, and zinc finger protein 91 (ZFP91) was detected in tissues and cells using quantitative real-time PCR. The protein level of ZFP91 was measured by Western blot analysis. Luciferase reporter, pull-down, and RNA immunoprecipitation assays were used to determine the relationship between MALAT1, miR-22-3p, and ZFP91. MTT assay was applied to measure cell survival and proliferation. Cell apoptosis was detected using flow cytometry. Tumor xenograft assay was used to detect the function of miR-22-3p in vivo.
Results: In this study, we found that MALAT1 and ZFP91 expression was upregulated while the expression of miR-22-3p was downregulated in GC/OXA tissues and cells. Additionally, miR-22-3p was a target miRNA of MALAT1 and ZFP91 was a target mRNA of miR-22-3p. Functional studies showed that the knockdown of MALAT1 or overexpression of miR-22-3p inhibited GC/OXA cell survival, proliferation, and drug resistance as well as induced apoptosis, which could be reversed by the inhibition of miR-22-3p or overexpression of ZFP91.
Conclusion: We observed a new regulatory network for MALAT1 in drug resistance of GC. MALAT1 modulates ZFP91 to promote GC cells OXA resistance via sponging miR-22-3p.
Keywords: GC, oxaliplatin, cell progression, MALAT1/miR-22-3p/ZFP91
Introduction
Gastric cancer (GC) is one of the most common cancers worldwide accompanied with high mortality and poor prognosis.1 Oxaliplatin (OXA) is a common medicine to treat various cancers including GC, belonging to the platinum-based antineoplastic family.2–4 However, some side effects have been observed following the treatment of cancer with OXA, such as ototoxicity, fatigue, nausea, vomiting, and rhabdomyolysis, which seriously affect patients’ treatment and prognosis.5–7 Therefore, a better understanding of the regulatory mechanism of OXA in GC can effectively improve the therapeutic effect of GC.
Recently, some studies have reported that lncRNAs play important roles in cell progression in many types of cancers, such as breast cancer, GC, and non-small cell lung cancer (NSCLC).8–10 Some lncRNAs are associated with cell proliferation, apoptosis, and drug-resistance in GC;11–13 eg, lncRNA HOXA11-AS contributed to the proliferation and invasion of GC cells.14 Upregulation of BANCR was implicated in clinical progression and poor prognosis.15 In addition, Wang et al reported that NEAT1 also reduced cell chemosensitivity in GC.16
miRNAs and lncRNAs belong to noncoding RNAs and increasing evidence demonstrated that lncRNA can act as competing endogenous RNAs (ceRNAs) to regulate mRNA by binding their common miRNAs in various cancers.17–20 These regulatory networks are widely involved in tumor occurrence, development, apoptosis, and drug resistance in GC. For instance, HOTAIR contributed to cisplatin resistance by regulating VEGFA and PIK3R2 via targeting miR-126 in GC.21 Interestingly, HOTAIR/miR-331-3p/HER2, SNHG5/miR-32/KLF4, and MT1JP/miR-92a-3p/FBXW7 axis were involved in GC cell proliferation, migration, and invasion.19–24
However, the relationship between ncRNAs and mRNA in GC under drug treatment has not been fully explored. In this study, we found that lncRNA MALAT1 was upregulated in GC and GC/OXA tissues and cells, suggesting that MALAT1 was closely associated with drug resistance in GC. To further explore the regulatory mechanism of MALAT1, we carried out a bioinformatic analysis and found that miR-22-3p was a potential target miRNA of MALAT1 and zinc finger protein 91 (ZFP91) was a potential target mRNA of miR-22-3p. In addition, miR-22-3p expression was reduced and ZFP91 expression was increased in GC and GC/OXA tissues and cells. Thus, we speculated that MALAT1 might regulate the expression of ZFP91 by competitively binding miR-22-3p to affect GC OXA resistance.
Materials and methods
Patients and tissues
Twenty four GC tissues and normal tumor-adjacent tissues were obtained from GC patients after informed written consent was obtained at Zhangye People’s Hospital Affiliated to Hexi University. Twenty four GC/OXA tissues were also obtained from GC patients whose resected tissues were confirmed by Response Evaluation Criteria in Solid Tumors at Zhangye People’s Hospital. The patients did not undergo any preoperation. This research was approved by the Research Ethics Committee of Zhangye People’s Hospital and followed the guidelines of the Declaration of Helsinki.
Cell culture and transfection
Normal cells (GES-1), GC cells (SGC-7901, BGC-823), and GC/OXA cells (SGC-7901/OXA and BCG-823/OXA, with lower OXA sensitivity than the corresponding GC cells) were purchased from RiboBio Co (Guangzhou, China) and all cells were cultured at 37°C with 5% CO2 in DMEM containing 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA), 1% penicillin, and 1% streptomycin.
siMALAT1, pcDNA-MALAT1 (MALAT1), pcDNA-ZFP91 (ZFP91), their negative control (NC) scramble, and pcDNA (vector) were obtained from GenePharma (Shanghai, China). In addition, miR-22-3p, anti-miR-22-3p, and their NC (miR-NC, anti-NC) were also obtained from GenePharma. All plasmids and fragments were transfected into SGC-7901/OXA and BCG-823/OXA cells using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Quantitative real-time PCR
TRIzol reagent (Thermo Fisher Scientific) was used to extract total RNAs from tissues and cells. cDNA was reverse-transcribed using TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) for miRNA and Reverse Transcription Reagents (Applied Biosystems) for mRNA and lncRNA. Quantitative real-time (qRT)-PCR was performed using SYBR Green (Takara Bio Inc, Japan). Glyceraldehyde phosphate dehydrogenase (GAPDH) and U6 were employed as internal control for mRNA/lncRNA and miRNA, respectively. 2−ΔΔCt method was used to detect the relative expression of mRNA/lncRNA and miRNA. The sequence of primers used was: lncRNA MALAT1, forward, 5′-AAAGTCCGCCATTTTGCCAC-3′ and reverse, 5′-ATTCGGGGCTCTGTAGTCCT-3′; ZFP91, forward, 5′-AGCTACCATTTGCCTACAA-3′ and reverse, 5′-GGGAAACGGCTGAGATAGTTT-3′; GAPDH, forward, 5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse, 5′-ATGGCATGGACTGTGGTCAT-3′; miR-22-3p, forward, 5′-AAGCTGCCAGTTGAAGAACTGTA-3′ and reverse, 5′-GCTGTCAACGATACGCTACGTAAC-3′; and U6, forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Western blot
The cells were lysed in RIPA buffer (Thermo Fisher Scientific). Total protein was separated using SDS-PAGE and transferred to polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). Then, the membrane was incubated with primary antibodies against ZFP91 and GAPDH (1:2,000; Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight. After incubation with secondary antibodies goat anti-mouse IgG (1:2,500; Santa Cruz Biotechnology Inc, Dallas, TX, USA) at 37°C for 1 hour, the blots were detected using an ECL detection kit (Thermo Fisher Scientific), and ODs were measured using ChemiDoc XRS System (Bio-Rad Laboratories Inc, Hercules, CA, USA).
Cell survival, proliferation, and apoptosis
MTT assay was used to measure cell survival and proliferation. Here, 2×103 cells were seeded into 96-well plates after treatment or transfection. After incubation for 48 hours, 20 μL MTT solution (5 mg/mL; Sigma-Aldrich Co, St Louis, MO, USA) was added into each well. Thereafter, 4 hours later, 200 µL dimethyl sulfoxide (Sigma-Aldrich Co) was added into each well. The absorbance at 490 nm was detected to measure cell survival and proliferation.
Flow cytometry (BD Biosciences, San Jose, CA, USA) was performed to assess cell apoptosis. After washing in PBS, the trypsinized cell was stained with Annexin V-FITC and propidium iodide (BD Biosciences) for 30 minutes.
RNA pull-down and RNA immunoprecipitation (RIP)
miR-22-3p and its NC were biotin-labeled with the Biotin RNA Labeling Mix (Hoffman-La Roche, Basel, Switzerland) and T7 RNA polymerase (Hoffman-La Roche) to the protein compound; then, the protein compound was purified using an RNasy Mini Kit (Qiagen NV, Venlo, the Netherlands). The SGC-7901/OXA cells were incubated with the protein compound at 25°C for 1 hour. Subsequently, qRT-PCR was used to evaluate the MALAT1 expression.
RIP was applied to further confirm the direct binding between MALAT1 and miR-22-3p or miR-22-3p and ZFP91 at the endogenous level in vitro using the EZ-Magna RIP kit (EMD Millipore), according to the manufacturer’s protocol. The SGC-7901/OXA cells were transfected with miR-22-3p and miR-NC for 48 hours and then incubated with an anti- Ago2 antibody and normal mouse IgG (EMD Millipore). Samples were incubated with Proteinase K with shaking to digest the protein. qRT-PCR was used to detect the expression of MALAT1 and ZFP91.
Luciferase reporter assay
The luciferase reporter vectors were constructed to contain 3′UTR of MALAT1 including wild-type (MALAT1-wt), mutant 3′UTR of MALAT1 (MALAT1-mut), 3′UTR of ZFP91 (ZFP91-wt), and mutant 3′UTR of ZFP91 (ZFP91- mut). HEK293T cells were seeded into a 96-well plate and were co-transfected with NC, miR-22-3p, anti-NC or anti-miR-22-3p, MALAT1-wt, MALAT1-mut, ZFP91-wt or ZFP91-mut using Lipofectamine 2000. After transfection for 48 hours, luciferase activity was detected with the Dual- Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA).
Animal experiments
BALB/c nude mice (4- to 6-week-old) were purchased from Vital River Laboratory Animal Technology Co, Ltd (Beijing, China) and maintained under controlled temperature (28°C) and specific pathogen-free conditions. The animal experiments were approved by the Institutional Animal Care and Use Committee of Zhangye People’s Hospital and carried out according to the National Institutes of Health guidelines for the care and use of laboratory animals. SGC-7901/OXA and BGC-823/OXA cells (1×107) transfected with miR-22-3p and their negative control (miR-NC) were injected subcutaneously into the right flank of the nude mice (n=5 per group). Six days later, the mice were intraperitoneally administrated with 2 mg/kg OXA every 5 days for five cycles. Tumor volumes were measured every 5 days until 25 days and were calculated using the formula: volume = length × width2/2. All mice were sacrificed to collect tumor xenografts and tumor xenografts were weighted.
Statistical analysis
All data are expressed as mean ± SD. Student’s t-test and one-way ANOVA were performed to analyze significant differences between the groups. Pearson’s correlation analysis was used to analyze the relationship between MALAT1 and ZFP91. GraphPad Prism 7.0 (GraphPad Software Inc, La Jolla, CA, USA) was used to display the analysis of results. P-values <0.05 were regarded as statistically significant.
Results
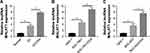
MALAT1 expression was increased in GC and GC/OXA tissues and cells
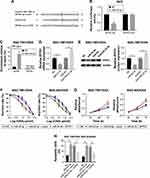
lncRNA has been demonstrated to play an important role in GC. Firstly, we checked the lncRNA MALAT1 expression in GC tissues and found that MALAT1 was significantly upregulated in GC tissues, when compared with normal tissues. Meanwhile, in GC/OXA tissues, MALAT1 expression was remarkably higher than that in GC tissues (Figure 1A). Next, we tested the MALAT1 expression in GC cell lines. As shown in Figure 1B and C, the expression of MALAT1 in SGC-7901 cells and BCG-823 cells was significantly higher than that in GES-1 cells, a normal cell line, whereas it was obviously lower than that in SGC-7901/OXA cells and BCG-823/OXA cells. Thus, these results suggested that MALAT1 may be associated with GC progression and drug resistance.
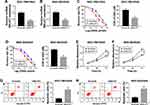
Knockdown of MALAT1 inhibited cell survival and proliferation as well as induced apoptosis in GC/OXA cells
To explore the function of MALAT1 in GC progression, we transfected siMALAT1 into SGC-7901/OXA and BCG-823/OXA cells to knockdown MALAT1. As shown in Figure 2A and B, siMALAT1 transfection obviously inhibited the expression of MALAT1 in SGC-7901/OXA and BCG-823/OXA cells, and reduced the IC50 of OXA. Next, we treated siMALAT1 and the scramble groups with different concentrations of OXA, and the results showed that the cell survival rate of siMALAT1 group was significantly lower than that of the scramble group (Figure 2C and D). Meanwhile, the MTT assay showed that siMALAT1 transfection significantly suppressed cell proliferation in SGC-7901/OXA and BCG-823/OXA cells (Figure 2E and F). Besides, the flow cytometry assay suggested that MALAT1 knockdown notably increased cell apoptosis in SGC-7901/OXA and BCG-823/OXA cells (Figure 2G and H). These findings suggested that MALAT1 knockdown could decrease GC/OXA cells survival, proliferation, and drug resistance as well as induce apoptosis.
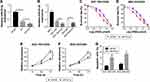
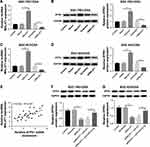
Overexpression of miR-22-3p inhibited cell survival and proliferation as well as induced apoptosis in GC/OXA cells
We also found that miR-22-3p was significantly decreased in GC tissues and cells compared with normal tissues and cells (Figure 3A). In addition, miR-22-3p expression was obviously decreased in GC/OXA cells (Figure 3B). Functional studies showed that overexpression of miR-22-5p inhibited GC/OXA cell survival (Figure 3C and D) and proliferation (Figure 3E and F) as well as promoted apoptosis (Figure 3G).
Suppressive effect of siMALAT1 on cell progression reversed by inhibition of miR-22-3p in GC/OXA cells
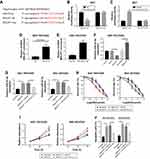
We predicted the relationship between miR-22-3p and MALAT1 using StartBase 2.0 (http://starbase.sysu.edu.cn/) and found that miR-22-3p has complementary sites with MALAT1 (Figure 4A). To confirm the binding relationship of MALAT1 and miR-22-3p, miR-22-3p, anti-miR-22-3p, and their negative control (NC and anti-NC) were co-transfected with MALAT1-wt or MALAT1-mut into HEK293T cells, respectively. Luciferase reporter assay showed that overexpression of miR-22-3p decreased the luciferase activity of MALAT1-wt and anti-miR-22-3p increased the luciferase activity of MALAT1-wt, but not MALAT1-mut (Figure 4B and C). More importantly, the RIP and pull-down assay further verified that miR-22-3p directly bound to MALAT1 (Figure 4D and E). These finding suggested that miR-22-ap was a target miRNA of MALAT1.
To further explore the function of MALAT1/miR-22-5p in GC/OXA cells, we detected the expression of miR-22-3p in control, vector, MALAT1, scramble, and siMALAT1 groups and found that the expression of miR-22-3p was significantly decreased after MALAT1 transfection and increased after siMALAT1 transfection (Figure 4F). Next, we divided the cells into four groups: scramble, siMALAT1, siMALAT1 + anti-miR-NC, and siMALAT1 + anti-miR- 22-3p. The expression of miR-22-3p was measured in these four groups (Figure 4G). Then, the survival rates (Figure 4H), proliferation (Figure 4I), and apoptosis (Figure 4J) were analyzed in these four groups after OXA treatment.
In conclusion, these effects were reversed by the inhibition of miR-22-3p in SGC-7901/OXA and BGC-823/OXA cells (Figure 4G–J). Therefore, the results demonstrated that miR-22-3p was a target of MALAT1 and MALAT1 affected GC cells through miR-22-3p.
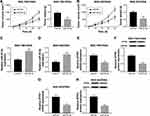
Overexpression of miR-22-3p inhibited cell growth and promoted apoptosis via inhibiting ZFP91 in GC/OXA cells
Considering that miRNA often play critical roles by targeting specific mRNA, we found that ZFP9 was a potential target mRNA using TargetScan (Figure 5A). As shown in Figure 5B, analysis of the luciferase reporter assay showed that luciferase activity was significantly decreased when miR-22-3p bound to ZFP91-wt, but not with ZFP91-mut (Figure 5B). RIP assay also determined that miR-22-3p directly targets ZFP91 (Figure 5C). Additionally, overexpression of miR-22-3p could inhibit the mRNA and protein expression level of ZFP91, whereas knockdown of miR-22-3p increased ZFP91 expression (Figure 5D and E). Following experiments suggested that overexpression of miR-22-3p suppressed cell survival and proliferation and induced apoptosis in SGC-7901/OXA and BGC-823/OXA cells, which was partially reversed by overexpression of ZFP91 (Figure 5F–H). Taken together, ZFP91 was a target mRNA of miR-22-3p and miR-22-3p inhibitor could inhibit GC/OXA cells growth and drug resistance, which could be reversed by upregulating ZFB91.
MALAT1 regulated ZFP91 through targeting miR-22-3p in GC/OXA cells
To clarify the relationship between MALAT1 and ZFP91, we observed GC cells with MALAT1 that were overexpressed and knocked down (Figure 6A–C). Then, we detected the protein expression level of ZFP91 in control, vector, MALAT1, scramble, and siMALAT1 groups. Western blot assay showed that the protein expression level of ZFP91 was promoted by overexpression of MALAT1, while it was inhibited by downregulation of MALAT1 in SGC-7901/OXA and BGC-823/OXA cells (Figure 6B–D). Meanwhile, the correlation analysis confirmed that MALAT1 was positively correlated with ZFP91 in GC/OXA tissues (Figure 6E). Besides, siMALAT1 decreased ZFP91 expression, which was impaired by inhibition of miR-22-3p in SGC-7901/OXA and BGC-823/OXA cells (Figure 6F and G), suggesting that MALAT1 regulated ZFP91 expression through modulating miR-22-3p.
miR-22-3p overexpression repressed tumor growth by regulating ZFP91 in vivo
To further verify the function of miR-22-3p in tumor growth, we transfected miR-22-3p and miR-NC into SGC-7901/OXA and BGC-823/OXA cells, respectively; then, the transfected cells were subcutaneously injected in the flank of nude mice. The results showed that promotion of miR-22-3p decreased tumor volume and weight in vivo (Figure 7A and B). More importantly, miR-22-3p transfection remarkably elevated miR-22-3p expression and reduced the mRNA and protein expression of ZFP91 (Figure 7C–H). Thus, upregulation of miR-22-3p significantly inhibited tumor growth in vivo.
Discussion
At present, the regulatory network of ncRNA/mRNA has been reported in many studies to play an irreplaceable role in drug resistance. As the upstream RNA of the regulatory network, lncRNAs play a vital role in many cancers. Accumulating evidence has shown that lncRNA MALAT1, a key regulatory factor, is associated with cell progression, epithelial–mesenchymal transition (EMT), and prognosis in cancer.25–27 MALAT1 was upregulated in many cancers, including GC, breast cancer, bladder cancer, and colorectal cancer.27–30 For example, upregulation of MTLAT1 improved migration of bladder cancer cells through promoting EMT by activating the Wnt signaling pathway in vitro.28 In addition, MALAT1 acts as a ceRNA participating in cancer progression and chemoresistance in GC. Li et al pointed out that MALAT1 modulated GC progression by regulating HMGB2 via competitively binding to miR-1297.31 MALAT1 modulated autophagy-associated chemoresistance through targeting miR-23b-3p in GC.32 In the present study, we found that MALAT1 is highly expressed in GC tissues and cells. Functional studies showed that low expression of MALAT1 significantly inhibited the viability and proliferation of GC/OXA cells and promoted apoptosis. These results indicated that lack of MALAT1 expression can reduce resistance in GC cells.
Many studies have shown that miR-22-3p is closely related to the cellular processes and chemoresistance in cancers, such as hepatocellular carcinoma, NSCLC, pancreatic cancer, colon cancer, and GC.33–36 Chang et al reported that miR-22-3p functions as a tumor inhibitor and suppresses cell proliferation by targeting AEG-1 in NSCLC.34 Also, in pancreatic cancer, plasma miR-22-3p, miR-642b-3p, and miR-885-5 serve as reliable biomarkers for diagnosis. In GC, miR-22 expression was significantly decreased and inhibited tumor growth, cell proliferation, invasion, and metastasis of GC through targeting MMP14, Snail, and CD151.36,37 In p53- mutated colon cancer, upregulation of miR-22 reversed paclitaxel-induced chemoresistance by activating PTEN signaling.38 In the present study, we found that miR-22-3p expressed lower in GC/OXA tissues and cells than that in GC tissues and cells. Exogenous introduction of miR-22-3p significantly inhibited the growth of GC/OXA cells and promoted apoptosis, indicating that upregulation of miR-22-3p can reduce the resistance of GC cells. In addition, using the luciferase reporter, pull-down, and RIP assays, we confirmed that miR-22-3p is the target miRNA of MALAT1. The reversed experiments further determined that the suppressive effect of siMALAT on cell progression was reversed by knockdown of miR-22-3p in GC/OXA cells. These results reflected that MALAT1 regulates the resistance of GC/OXA cells by modulating miR-22-3p.
ZFP91 has been reported to play an important role in cancer progression and tumorigenesis, especially in GC.39,40 Peng et al showed that ZEP91 was a target of miR-188-5p and together with it regulated GC cell proliferation and invasion.41 In our study, ZFP91 was a target mRNA of miR-22-3p and overexpression of ZFP91 could alleviate the inhibitory effect of overexpression of miR-22-3p on GC/OXA cells. Besides, we also found that MALAT1 regulated ZFP91 expression by completely binding miR-22-3p in GC/OXA cells. Animal experiments showed that overexpression of miR-22-3p significantly suppressed GC/OXA tumor growth in vivo and reduced ZFP91 expression, revealing that miR-22-3p affects GC tumor drug resistance through directly regulating ZFP91 expression.
Conclusion
Knockdown of MALAT1 could inhibit GC/OXA cell growth and drug resistance as well as induce cell apoptosis through upregulation of miR-22-3p by inhibition of ZFP91, suggesting that the MALAT1/miR-22-3p/ZFP91 axis plays an important regulatory role in GC/OXA resistance.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2015;61(2):69–90. doi:10.3322/caac.20107
2. Apps MG, Choi EH, Wheate NJ. The state-of-play and future of platinum drugs. Endocr Relat Cancer. 2015;22(4):R219–R233. doi:10.1530/ERC-15-0237
3. Bang Y, Kim YW, Yang H, et al. Adjuvant capecitabine and oxaliplatin for gastric cancer: results of the phase III CLASSIC trial. J Clin Oncol. 2011;29(15_suppl):LBA4002. doi:10.1200/jco.2011.29.15_suppl.lba4002
4. André T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350(23):2343–2351. doi:10.1056/NEJMoa032709
5. Cassidy J, Misset JL. Oxaliplatin-related side effects: characteristics and management. Semin Oncol. 2002;29(5):11–20. doi:10.1053/sonc.2002.35524
6. Hijri FZ, Arifi S, Ouattassi N, Mellas N, El Mesbahi O. Oxaliplatin-induced ototoxicity in adjuvant setting for colorectal cancer: unusual side effect. J Gastrointest Cancer. 2014;45(1):106–108. doi:10.1007/s12029-013-9489-3
7. Oun R, Moussa YE, Wheate NJ. The side effects of platinum-based chemotherapy drugs: a review for chemists. Dalton Trans. 2018;47(19):6645–6653. doi:10.1039/C8DT00838H
8. Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6(13):11652–11663. doi:10.18632/oncotarget.3457
9. Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi:10.18632/oncotarget.1913
10. Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi:10.1016/j.canlet.2015.11.024
11. Fang Q, Chen XY, Zhi XT. Long non-coding RNA (LncRNA) Urothelial Carcinoma Associated 1 (UCA1) increases multi-drug resistance of gastric cancer via downregulating miR-27b. Med Sci Monit. 2016;22:3506–3513.
12. Zhang Y, Song X, Wang X, Hu J, Jiang L. Silencing of LncRNA HULC enhances chemotherapy induced apoptosis in human gastric cancer. J Med Biochem. 2016;35(2):137–143. doi:10.1515/jomb-2015-0016
13. Hu Y, Zhen M, He Y, Wei L, Yu S, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491(4):926–931. doi:10.1016/j.bbrc.2017.07.137
14. Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76(21):6299–6310. doi:10.1158/0008-5472.CAN-16-0356
15. Li L, Zhang L, Zhang Y, Zhou F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed Pharmacother. 2015;72:109–112. doi:10.1016/j.biopha.2015.04.007
16. Wang J, Niu Q, Shi N, et al. Long non-coding RNA NEAT1 decreases the chemosensitivity of gastric cancer cells via regulating P-glycoprotein expression. Mol Cell Toxicol. 2017;13(3):317–325. doi:10.1007/s13273-017-0035-4
17. Hao NB, He YF, Li XQ, Wang K, Wang RL. The role of miRNA and lncRNA in gastric cancer. Oncotarget. 2017;8(46):81572–81582. doi:10.18632/oncotarget.19197
18. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi:10.1158/2159-8290.CD-13-0202
19. Zhang G, Li S, Lu J, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17(1):87. doi:10.1186/s12943-018-0829-6
20. Chen Y, Shen Z, Zhi Y, et al. Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophys. 2018;645:117–125. doi:10.1016/j.abb.2018.03.018
21. Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biol. 2016;37(12):1–11. doi:10.1007/s13277-016-5448-5
22. Liu XH, Sun M, Nie FQ, et al. LncRNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13(1):92. doi:10.1186/1476-4598-13-92
23. Ni Z, Zhang D, Xie H, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351(1–2):157–164. doi:10.1007/s11010-011-0723-7
24. Zhao L, Han T, Li Y, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2016;31(3):893–903. doi:10.1096/fj.201600994R
25. Gutschner T, Hämmerle M, Eißmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi:10.1158/0008-5472.CAN-12-2850
26. Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011; 6(12):1984–1992. doi:10.1097/JTO.0b013e3182307eac
27. Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174.
28. Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8(9):2289–2294. doi:10.1039/c2mb25070e
29. Chou J, Wang B, Zheng T, et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472(1):262–269. doi:10.1016/j.bbrc.2016.02.102
30. Ying Q, Hong SO, Wu J, et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget. 2016; 7(11):12693–12703. doi:10.18632/oncotarget.7281
31. Li J, Gao J, Wen T, Li Y, Zhang J. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int. 2017;17(1):44. doi:10.1186/s12935-017-0408-8
32. Yiren H, Yingcong Y, Sunwu Y, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR- 23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16(1):174. doi:10.1186/s12943-017-0743-3
33. Chen J, Wu FX, Luo HL, et al. Berberine upregulates miR-22-3p to suppress hepatocellular carcinoma cell proliferation by targeting Sp1. Am J Transl Res. 2016;8(11):4932–4941.
34. Chang Y, Yan W, Jiang X, et al. MiR-22-3p suppresses cell proliferation by targeting AEG-1 in NSCLC. J Pract Med. 2017;14:2267–2271.
35. Hussein NAEM, Kholy ZAE, Anwar MM, Ahmad MA, Ahmad SM. Plasma miR-22-3p, miR-642b-3p and miR-885-5p as diagnostic biomarkers for pancreatic cancer. J Cancer Res Clin Oncol. 2017; 143(1):83–93. doi:10.1007/s00432-016-2248-7
36. Wang X, Yu H, Lu X, Zhang P, Wang M, Hu Y. MiR-22 suppresses the proliferation and invasion of gastric cancer cells by inhibiting CD151. Biochem Biophys Res Commun. 2014;445(1):175–179. doi:10.1016/j.bbrc.2014.01.160
37. Zuo QF, Cao LY, Yu T, et al. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis. 2015;6(11):e2000. doi:10.1038/cddis.2015.297
38. Li J, Zhang Y, Zhao J, Kong F, Chen Y. Overexpression of miR-22 reverses paclitaxel-induced chemoresistance through activation of PTEN signaling in p53-mutated colon cancer cells. Mol Cell Biochem. 2011;357(1–2):31–38. doi:10.1007/s11010-011-0872-8
39. Xu SH, Zhu S, Wang Y, et al. ECD promotes gastric cancer metastasis by blocking E3 ligase ZFP91-mediated hnRNP F ubiquitination and degradation. Cell Death Dis. 2018;9(5):479. doi:10.1038/s41419-018-0525-x
40. Ma J, Mi C, Wang KS, Lee JJ, Jin X. Zinc finger protein 91 (ZFP91) activates HIF-1αviaNF-κB/p65 to promote proliferation and tumorigenesis of colon cancer. Oncotarget. 2016;7(24):36551–36562. doi:10.18632/oncotarget.9070
41. Peng Y, Shen X, Jiang H, et al. MiR-188-5p suppresses gastric cancer cell proliferation and invasion via targeting ZFP91. Oncol Res. 2018; 27:65–71. doi:10.3727/096504018X15191223015016
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.