Back to Journals » OncoTargets and Therapy » Volume 13
lncRNA LSINCT5 Regulates miR-20a-5p/XIAP to Inhibit the Growth and Metastasis of Osteosarcoma Cells
Authors Liao SA, Guan J, Mo H, He JL, Zhan XL
Received 28 February 2020
Accepted for publication 24 June 2020
Published 18 August 2020 Volume 2020:13 Pages 8209—8221
DOI https://doi.org/10.2147/OTT.S251843
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
This paper has been retracted.
Shi-An Liao, 1 Jian Guan, 2 Hao Mo, 2 Ju-Liang He, 2 Xin-Li Zhan 1
1Department of Spine and Osteopathy Ward, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Province, People’s Republic of China; 2Department of Bone and Soft Tissue Surgery, Guangxi Medical University Cancer Hospital, Nanning, Guangxi Province, People’s Republic of China
Correspondence: Xin-Li Zhan
Department of Spine and Osteopathy Ward, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi 530021, People’s Republic of China
Tel +86-13907864389
Email [email protected]
Background: More and more evidence has shown that non-coding RNA (ncRNA), including long ncRNA (lncRNA) and micro RNA (miRNA), plays a crucial regulatory role in osteosarcoma (OS). Previously, we revealed a Rho-related coiled coil incorporating protein kinase 1(XIAP). A transfer-related gene is negatively regulated by microRNA-20a-5p (miR-20a-5p) and plays the role of oncogene in OS. It is not clear if any lncRNA is involved in the axial upstream of miR-20a-5p/XIAP.
Methods: Expression of LSINCT5 and miR-20a-5p/XIAP in OS tissues was determined through qRT-PCR (qP). The proliferation and migration/invasion activity of OS cells were tested through CCK-8/and transwell assay, respectively. The changes on expression of XIAP were examined through qRT-PCR and Western blot (WB). Targeted binding between LSINCT5, miR-20a-5p, and XIAP has been verified using dual luciferase reporter gene analysis, RNA Immunoprecipitation (RIP), and RNA pull-down experiments. The effect of LSINCT5 on tumor growth was determined by tumor allograft test.
Results: In this study, elevated LSINCT5 was found in OS tissue samples and OS cell strains, and the increased LSINCT5 was strongly related to the adverse prognosis of clinical patients. Functional assays showed that inhibition of LSINCT5 could up-regulate miR-20a-5p-mediated OS cells proliferation and metastasis. WB analysis and qP analysis showed that LSINCT5 regulated XIAP by mediating miR-20a-5p. Further cell behavior experiments showed that LSINCT5 acted as a miR-20a-5p sponge to inhibit proliferation and metastasis caused by XIAP. Finally, the results of animal models in vivo showed that LSINCT5 could regulate the tumor growth of OS.
Conclusion: LncRNA LSINCT5 acts as an oncogene and promotes XIAP mediated growth and metastasis as competitive endogenous RNA (ceRNA) in OS.
Keywords: lncRNA, LSINCT5, miR-20a-5p, osteosarcoma, growth, invasion
Introduction
Osteosarcoma (OS) is one of the most common primary bone malignant tumors in children and adolescents.1 In China’s cancer epidemiology statistics in 2015,2 there wee 28,000 newly diagnosed patients with OS and 20,700 OS-related deaths. At present, the main clinical treatment of OS is mainly surgery, but in most cases, the tumor of patients diagnosed with OS has transferred to the lungs, which greatly reduces the effectiveness of treatment and affects the prognosis of patients.3,5 Although the prognosis of some patients can be improved through radiotherapy and chemotherapy, long-term drugs can easily lead to drug resistance and tumor relapse, and lower the survival of patients for the long term.6 Thus, it is a must to better understand the pathogenesis and molecular mechanism of OS, which will contribute to the clinical treatment and prognosis of OS.
lncRNA is a non-coding RNA with a length of more than 200 nt.7 Previously, lncRNA had no protein coding function and was considered as “noise” generated by the transcription process. However, recent studies have found that lncRNA has differential expression in various diseases.8,9 Among them, in tumor-related studies, lncRNA plays a key role in the diagnosis and prognosis of tumors. For example, in the research of Li et al,10 four lncRNA linked to the prognosis of breast carcinoma were found through the analysis of the lncRNA co-expression network. Another study found that lncRNA MVIH has a high value in the prognosis and clinical pathology of cancer patients.11 Long stress-induced non-coding transcript 5 (LSINCT5) is a newly discovered lncRNA in recent years. Previous studies have found that LSINCT5 can be used as a prognostic indicator for OS and plays a vital part in tumor carcinogenesis.12 Nevertheless, the relevant mechanism of LSINCT5 in OS is still unclear.
Through online prediction, we concluded that LSINCT5 and miR-20a-5p had a targeted binding locus. Early studies have uncovered that miR-20a-5p is weakly expression in OS and it is a potential therapeutic target for OS. Thus, this research was designed to seek the mechanism of LSINCT5 and miR-20a-5p in OS and provide potential targets for clinical use.
Methods and Materials
Collection of Patients’ Samples
From May 2012 to May 2014, 80 patients with OS treated in our hospital were collected. Carcinoma tissues and para-carcinoma tissues of patients were obtained during the operation, transported with fluid nitrogen, and stored at −80°C. The patients had not received anti-tumor treatment before this study. All patients cooperated with follow-up. This test was ratified by the Medical Ethics Committee of our Guangxi Medical University Cancer Hospital. All patients were informed about this study and signed written informed consent. The study was conducted in accordance with the Helsinki Declaration.
Cells Culturing
Human OS cells SOSP-9607, MG-63, U2OS, SAOS-2, and bone cell line (hFOB) from American type culture collection center (ATCC) were cultivated by DMEM (Dulbecco modified Eagle medium), which contained 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) and 100 U/mL penicillin/streptomycin. The cells were cultivated at 37°C and 5% CO2.
Cells Transfection
The specific short hairpin RNA (shRNA) directed against human lncRNA LSINCT5 was cloned into pENTR TM/U6 plasmid (GenePharma, Shanghai, China) and called sh-LSINCT5. Non-targeted shRNA (sh-NC, GenePharma) was applied as a negative control. The full-length sequence of lncRNA LSINCT5 was transfected into pcDNA-3.1 vector (ThermoFisher Scientific, China) and called pcDNA-LSINCT5. An empty pcDNA vector was used as a negative control. miR-20a-5p mimics (miR-20a-5p-mimics), inhibitors (miR-20a-5p-inhibit), or corresponding perturbation controls (miR-NC) were synthesized by RiboBio (Guangzhou, China). XIAP-specific siRNA (si-XIAP) and siRNA negative pair (si-NC) were from Santa Cruz (United States). For overexpression of XIAP, the full-length XIAP sequence was transfected into a pDNA-3.1 vector (ThermoFisher Scientific) and called pcDNA-XIAP. pDNA-3.1 was use as blank control. On the basis of the manufacturer’s plan, all transfection was processed by Lipofectamine 3000 reagent. Stable transfected MG-63 and U2OS cells were selected by a medium incorporating 0.5 mg/mL G418 (Sigma-Aldrich, St. Louis, MO, USA). Stable transfected cells were selected for succeeding tests.
Detection of Cells Proliferation (CCK-8)
The CCK-8 kit was used to test cell proliferation. The specific detection steps were as follows: transfected MG-63 and U2OS cells were cultivated in 96-well plates, and cultivated for 24, 48, and 72 hours after transfection. CCK 8 analysis was conducted to test cell proliferation. The absorbance at 450 nm was tested on the enzyme-labeling instrument (the United States).
Cells Invasion and Migration
The BD Matrigel chamber (BD Biosciences, UK) was used to detect cell invasion. The specific detection steps were as follows: transfected MG-63 and U2OS cells were inoculated into a membrane chamber in a serum-free culture medium, and the medium incorporating 10% FBS was added to the bottom chamber. After 24 hours, the cells in the dark room were dyed through crystal violet (Sigma-Aldrich, St. Louis, MO, USA) and counted. Cell migration was tested by an improved two-chamber migration analysis chamber of 8 μm polycarbonate membrane (Costar-Corning, New York, USA). The specific detection steps were as follows: transfected MG-63 and U2OS cells were suspended in 200 μL medium without serum and inoculated in the upper compartment of the 24-well chamber. Complete medium (600 μL) was put in the lower compartment. After hatch for 12 hours, it was fixed with methanol for 30 minutes, dyed through 0.1% crystal violet (Amresco, USA), and calculated under a microscope.
Flow Cytometry
The transfected MG-63 and U2OS cells were detected for apoptosis by Annexin V-PE cell apoptosis detecting kit. For single cell suspensions, it was necessary to digest cells with trypsin. Then, what we needed to do was wash the cells with cold PBS and resuspend them in binding buffer. The concentration was adjusted to 1×106 cells/mL. After that, the suspension of cells was absorbed by about 100 μL. Then, the cell suspension was placed in an inflow tube (5 mL) and then incubated with Annexin V-FITC (5 µL, BioVision, Milpitas, CA) and PI (10 µL, 20 µg/L, PI, Sigma-Aldrich). Next, they were placed in the dark at room temperature for 15 minutes. PBS (400 µL) was added to the reaction tube. FACS Calibur was then used for apoptosis. FACS Diva was used to analyze data. The experiment was repeated three times.
qPCR (RT-qPCR)
According to the manufacturer’s regulations, TRIzol (Invitrogen) was applied to extract total RNA from cultured cells or tissues. SYBR (Takara, China) and qRT-PCR were applied to quantify lncRNA, microRNA (miR) and mRNA on 7900HT system. The reaction system and reaction regulation were carried out according to the kit instructions. mRNA and lncRNA used GAPDH as an internal reference. miR used U6 as an internal reference. The relative expression value was analyzed by the (2−ΔΔCT).13
WB Assay
RIPA buffer was applied to obtain the total protein of cells. BCA (Thermo, PA, USA) was applied to prepare and test the total protein of cells. The total protein was isolated on 12% SDS-PAGE and moved to PVDF membrane. The membrane was sealed through dry milk and the total protein was immunostained with primary antibody X-linked inhibitor of apoptosis (XIAP) and GAPDH at −4°C for 1 night. After incubation with a second antibody, the signal was visualized through the chemiluminescence testing system (Pierce, ThermoA).
Double Fluorescein Report
The following four pmiR-RB-REPORT TM vectors were synthesized: XIAP 3ʹ-UTR, containing miR-20a-5p putative target loci (XIAP WT-3ʹ-UTR); XIAP 3ʹ-UTR (XIAP Mut-3ʹ-UTR) with mutation binding site; Full-length LSINCT5, including the putative target of miR-20a-5p (LSINCT5-WT); Full-length LSINCT5 containing mutation combining site (LSINCT5-Mut). In total, 100 ng of vector (XIAP WT-3ʹ-UTR, XIAP Mut-3ʹ-UTR, LSINCT5-WT, or LSINCT5-Mut) and miR-20a-5p mimics or mimetic control (50 nM/well) were transfected into 293T and SW1353 cells by riboFECT TM CPRibobio reagent. Analog control and XIAP Mut-3ʹUTR were applied as NC. Fluorescein activity was tested through a dual luciferase reporter kit (Promega, Madison, WI, USA).
RIP
RIP detection was performed through a Magna RNA binding protein immunoprecipitation kit. It was as follows: Whole cell lysate was cultivated through RIP buffer incorporating magnetic beads coupled to human anti-Ago2 antibody or normal mouse IgG as negative control. The sample was cultivated with proteinase K, and then immunoprecipitated RNA was segregated. The immunoprecipitated RNA was purified and then analyzed by qP to quantify LSINCT5 and miR-20a-5p.
RNA Pull-Down Experiment
1RNAg biotin-labeled RNA was put in Eppendorf (EP) tubes by magnetic RNA-protein pull-down kit. μ-Protein Pull-Down Kit (Pierce, Rochford, IL, USA) was used. Then 500 μL of structural buffer was added, and a water bath was conducted at 95°C for 2 minutes. The magnetic beads were completely resuspended, and then 50 μL of the magnetic bead suspension was put in the EP tube at 4°C overnight and centrifuged at 3000 rpm for 3 minutes. Then, the supernatant was removed. After adding 500 μL RIP washing buffer 3 times, beads, and 10 μL cell lysate were put and laid up at ambient temperature for 1 hour. The cultured magnetic bead–RNA-protein complex was centrifuged at low velocity. The supernatant was obtained and rinsed 3 times with 500 μL RIP washing buffer on the basis of the manufacturer’s specifications. Cell lysate supernatant of 10 μL was employed to quantify miR-20a-5p.
RNA-FISH
FISH analysis was performed using Ribo™ fluorescence in situ cross kit (Ribobio Company, China). LSINCT5 and U6 probes were designed and synthesized by Ribobio Company and labeled with Cy3 fluorescent dye. According to the manufacturer’s instructions, fluorescence in situ cross kit was used for fluorescence detection with a confocal laser scanning microscope (Leica, Germany).
Nude Mouse Model in vivo
There were 15 BALA/C nude mice (4 weeks old, weight of 18–25 g, male, Beijing Weitong Lihua Company). Nude mice were subcutaneously injected with stable expression of lentivirus (sh-NC), overexpression of LSINCT5 lentivirus (pcDNA-LSINCT5), and inhibition of expression of LSINCT5 lentivirus (sh-LSINCT5) in U2OS cells (1×105/cells). The mice were subcutaneously inoculated into the right posterior dermis (each n=5). The tumor size was evaluated and computed on the basis of the following equation (volume=(maximum diameter×minimum diameter2)/2, unit=mm3). Mice were euthanized by neck dislocation method 28 days after injection. The tumor tissues were taken out and weighed. All animal experiments had been approved by the Ethics Committee of Guangxi Medical University Cancer Hospital. The animal experiment guide referred to the “Laboratory animal—Guideline for ethical review of animal welfare” (GB/T 35,892–2018) issued by China in 2018.
Statistical Analysis
GraphPad 7 was applied to analyze the data. The independent sample t-test was used for inter-group comparison. The counting data were expressed as a percentage (%). The chi-square test was expressed by χ2. One-way ANOVA was used for multi-group comparison, expressed as F. LSD t-test was used for pairwise comparison afterwards. Repetitive measurement and analysis of variance was used for expression at multiple time points, expressed as F. Bonferroni was used for post test. Pearson test was applied to analyze the correlation of each gene. The K-M survival curve was used to draw the total survival condition of patients. Log rank test was applied for analysis. There was a statistical difference with P<0.05.
Results
The Expression of LSINCT5 in OS Increased and the Survival Rate of Patients Decreased
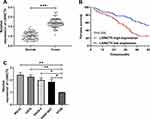
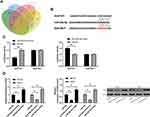
To determine the expression of LSINCT5 in OS, we tested the relative expression of LSINCT5 in tumor tissues of patients with OS. These results revealed that the expression of LSINCT5 in OS cancer tissues increased (Figure 1A), and it was also concluded that patients with high LSINCT5 expression showed high Enneking stage staging, and the probability of distal metastasis was significantly increased (Table 1). In addition, the 5-year survival rate was obviously reduced in patients with high LSINCT5 expression after follow-up (Figure 1B). Through the detection of OS cells, we found that the expression of LSINCT5 in OS was obviously enhanced (Figure 1C). These studies suggested that LSINCT5 could be used as a potential target for the treatment of OS.
 |
Table 1 Relationship Between LSINCT5 and Pathological Data of Osteosarcoma Patients |
Effect of LSINCT5 on Growth and Metastasis of OS
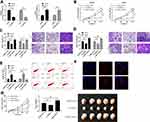
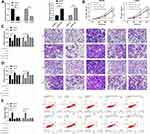
In order to further seek the impact of LSINCT5 on the growth of OS cells, we selected OS cells with significant expression to perform LSINCT5 knock-down, and observed the effect on OS cells after LSINCT5 knock-down (Figure 2A). CCK-8 test showed that after LSINCT5 knock-down, the proliferation ability of OS cells was inhibited compared with OS cells transfected with sh-NC (Figure 2B). Transwell was used to detect the cell invasion and migration and showed that, after LSINCT5 knock-down, the number of cell membrane penetration and migration of OS cells was significantly inhibited compared with OS cells transfected with sh-NC (Figure 2C and D). However, flow cytometry showed that knocking down LSINCT5 induced apoptosis of OS cells, while transfection and injection of pcDNA-LSINCT5 reversed cell proliferation, invasion, migration, and apoptosis (Figure 2E). RNA-FISH showed that most positive cells were located in the cytoplasm and a few in the nucleus (Figure 2F). Besides, our research also revealed that injecting stable sh-LSINCT5 subcutaneously into nude mice effectively inhibited the growth of the tumor, while cell growth accelerated after injecting pcDNA-LSINCT5 (Figure 2G). This further revealed that LSINCT5 could be a potential target for treatment of OS.
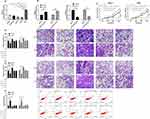
LSINCT5 Could Act as miR-20a-5p Sponge to Control the Growth of OS Cells
LcnRNA has been confirmed as the ceRNA (competing endogenous RNAs) of miR in various studies on tumor mechanisms. In order to explore that LSINCT5 could bind miR through miRDB for prediction,14 we concluded that miR-20a-5p and LSINCT5 had binding locus (Figure 3A). In order to verify the relationship between the two, we carried out an experiment to verify it. The double luciferase report detection found that miR-20a-5p-mimics could inhibit the fluorescence activity of LSINCT5-WT, while the fluorescence activity of LSINCT5-WT co-transfected with miR-20a-5p-inhibit was obviously up-regulated, which revealed that LSINCT5 could specifically bind with miR-20a-5p (Figure 3B). To further study their relationship, RIP experiments found that LSINCT5 and miR-20a-5p could bind to Ago2 protein, and the expressions of LSINCT5 and miR-20a-5p bound to Ago2 were higher than those of LSINCT5 and miR-20a-5p bound to immunoglobulin (Ig) G (Figure 3C). In addition, we also conducted RNA pull-down experiments. The results showed that the enrichment of miR-20a-5p enhanced in response to transfection with LSINCT5-WT, while the enrichment of miR-20a-5p changed in LSINCT5-MUT compared with Bio-NC (Figure 3D). Not only that, we also detected OS cells transfected with pcDNA-LSINCT5 and sh-LSINCT5. This result also revealed that expression of miR-20a-5p in cells transfected with pcDNA-LSINCT5 decreased, while expression of miR-20a-5p in cells transfected with sh-LSINCT5 reversed (Figure 3E). These experiments suggested that LSINCT5 could specifically bind to miR-20a-5p. For the purpose of studying their regulatory influence in cells more deeply, we conducted cell experiments and revealed that miR-20a-5p was low expressed in OS cells (Figure 4A and B). After up-regulating miR-20a-5p, the proliferation (Figure 4C), invasion, and migration (Figure 4D and E) of cells were obviously inhibited and apoptosis was induced (Figure 4F), while after down-regulating miR-20a-5p, the biological functions of cells were reversed. In addition, we detected the biological function of cells after co-transfection of miR-20a-5p-mimics with pcDNA-LSINCT5 and miR-20a-5p-inhibit with sh-LSINCT5. It was concluded that there was no obvious difference in cell biological behavior after co-transfection compared with miR-NC. Through the above research, we revealed that LSINCT5 could regulate the growth and metastasis of OS cells by specifically binding miR-20a-5p. More details are shown in Figures 3 and 4.
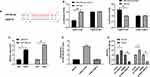
miR-20a-5p Targeted Regulation of XIAP to Inhibit the Growth of OS
miR has been proved by many studies to affect tumor growth by targeting downstream target genes. For the purpose of exploring the action of miR-20a-5p more deeply, we predicted its target genes through online prediction websites of Targetscan, miRDB, starBase, and miRTarBase (Figure 5A).15,17 We found that XIAP had a targeted relationship with miR-20a-5p in all four websites (Figure 5B). In order to conclude the targeting relationship between miR-20a-5p and XIAP, we carried out double luciferase activity detection. This result revealed that miR-20a-5p-mimics could inhibit XIAP-WT fluorescence activity, while XIAP-WT fluorescence activity co-transfected with miR-20a-5p-inhibit was obviously enhanced, which revealed that XIAP could be used as a target gene downstream of miR-20a-5p (Figure 5C). Moreover, we also detected the relative mRNA and protein expression of XIAP in OS cells transfected with miR-20a-5p-mimics and miR-20a-5p-inhibit. This result revealed that the relative mRNA and protein expression of XIAP in cells was obviously inhibited after enhancement of miR-20a-5p, while the results were reversed after down-regulation of miR-20a-5p (Figure 5D). This researchrevealed that miR-20a-5p could target XIAP. In order to further observe the impact of XIAP on the growth of OS cells, we transferred different XIAP expression vectors (si-XIAP, pcDNA-XIAP) into OS cells (Figure 6A). Through observation, we found that the growth (Figure 6B), migration (Figure 6D), and invasion (Figure 6C) of OS cells were inhibited after XIAP knockdown, and apoptosis was further induced (Figure 6E), while the up-regulation of XIAP facilitated the growth, invasion, migration of OS cells, and controlled the apoptosis, which indicated that XIAP participated in the growth and metastasis of OS cells. Besides, we observed the influence of si-XIAP and miR-20a-5p-inhibit and co-transfection of pcDNA-XIAP with miR-20a-5p-mimics on cell growth and transfection through co-transfection. The cell proliferation, invasion, migration, and apoptosis after co-transfection had no difference compared with si-NC+pcDNA-3.1, which suggested that miR-20a-5p can target and regulate XIAP to inhibit OS growth.
Expression of miR-20a-5p and XIAP in Patients’ Tissues and Correlation Analysis with LSINCT5
At the end of the research, we further detected the miR-20a-5p and XIAP in the cancer tissue of OS patients. RT-qPCR detection revealed that the miR-20a-5p in the cancer tissue of OS patients was obviously reduced (Figure 7A), while the expression of XIAP was obviously enhanced (Figure 7B). Further correlation analysis showed that LSINCT5 expression in tumor tissue of OS patients was negatively related to miR-20a-5p and positively related to XIAP (Figure 7C). We also found that XIAP and miR-20a-5p in tumor tissue of OS patients were negatively correlated (Figure 7C), which revealed the regulatory relationship between LSINCT5 and miR-20a-5p/XIAP axis from the side.
Discussion
OS is a common orthopedic tumor in clinical practice. Although the treatment plan for OS is constantly improved, the prognosis and survival of OS patients are still an important problem at present.18 Therefore, it is particularly important to explore the mechanism of OS for providing potential therapy targets for clinic and improving the prognosis of patients.
Recent studies have found that lncRNA plays an important role in the formation and development of various diseases.19 As a newly discovered lncRNA in recent years, LSINCT5 is located on the human 5p15.33 chromosome. Previous research has shown that LSINCT5 affects the development and progression of colorectal carcinoma, ovarian cancer,20 gastric cancer,21 esophageal cancer, and chronic heart failure,22,23 but there is relatively little research on OS at present. However, our study revealed that LSINCT5 was highly expressed in OS tissues and cell strains, and the 5-year survival of patients with high expression decreased, which was consistent with the study of He et al.12 However, the relevant mechanism is still unclear. Therefore, we conducted tests to further seek the potential mechanism of LSINCT5 in OS.
First of all, to determine the impact of LSINCT5 on OS cells, we knocked down and increased the expression of LSINCT5 in OS cells, respectively. The result showed that the cell proliferation, invasion and migration after knocking down LSINCT5 were inhibited compared with the control, and cell apoptosis was further induced. However, observation of LSINCT5 OS cells with up-regulated transfection showed that the cell growth, invasion, and migration were accelerated, and the apoptosis rate was inhibited. The low survival rate of OS is mainly due to the fact that patients are prone to focus metastasis. Previous studies have shown that LSINCT5 can inhibit the metastasis of lung cancer by regulating HMGA2,24 while our research has found that LSINCT5 also has the function of inhibiting tumor cell metastasis in OS, which has indicated that LSINCT5 has the function of inhibiting tumor metastasis in various tumors. In order to further explore the mechanism of LSINCT5, we predicted the miR that LSINCT5 could bind to it.
The theory of ceRNA has accelerated the research on lncRNA.25 Most studies have revealed that lncRNA competed with miR for the original reaction,26,28 thus causing changes in miR transcription and expression. In this research, we used miRDB prediction to find that miR-20a-5p and LSINCT5 had binding locus. MiR-20a-5p is a common tumor suppressor gene. Early studies have uncovered that miR-20a-5p is expressed in gastric cancer,29 lung cancer, and breast cancer, and had certain diagnostic value.30,31 In addition, the studies by Pu et al32 and Zhao et al33 have shown that miR-20a-5p can target KIF26B and SDC2 to inhibit multi-drug resistance of OS, respectively. In order to conclude the relationship between LSINCT5 and miR-20a-5p, we concluded that LSINCT5 and miR-20a-5p could bind specifically through double luciferase report. RIP and RNA pull-down experiments both revealed that LSINCT5 could bind miR-20a-5p. Besides, the miR-20a-5p in OS cells after transfection of pcDNA-LSINCT5 and sh-LSINCT5 was also changed. In order to confirm that LSINCT5 can regulate miR-20a-5p to inhibit the growth and metastasis of OS cells, cell experiments were conducted and showed that the transfer and growth of cells transfected with miR-20a-5p-mimics were inhibited, but the transfer and growth of cells transfected with miR-20a-5p-inhibit were accelerated. Besides, the experiment revealed that there was no obvious difference in cell transfer and growth after co-transfection of miR-20a-5p-mimics with miR-20a-inhibit and sh-LSINCT5 with pcDNA-LSINCT5 compared with the control, which indicated that LSINCT5 could regulate miR-20a-5p to affect the growth and transfer of OS cells.
XIAP is one of the members of the family of inhibitors of apoptosis proteins, which plays a role by binding to tumor necrosis factor receptor-related factors TRAF1 and TRAF2.34,35 Previous studies have found that XIAP is highly expressed in various tumors and inhibited tumor cell apoptosis.36,37 In addition, studies by Liu et al38 and Zheng et al39 have shown that miR-377 and miR-320 can both target XIAP to inhibit multi-drug resistance of OS. However, we predicted the miR-20a-5p target gene and found that XIAP and miR-20a-5p had binding locus. Furthermore, we revealed the targeted relationship between the two through double luciferase report, and verified the XIAP mRNA and protein in OS cells transfected with miR-20a-5p-mimics and miR-20a-inhibit. To observe the regulating function of miR-20a-5p and XIAP, we co-transfected pcDNA-XIAP with miR-20a-5p-mimics and si-XIAP with miR-20a-inhibit according to the previous scheme. This results revealed that the growth, invasion, and migration of OS cells were accelerated and the apoptosis rate was decreased after transfection of pcDNA-XIAP, while the results were reversed after co-transfection with miR-20a-5p-mimics. si-XIAP and miR-20a-inhibit had similar situations.
At the end of the study, we also quantified tissue miR-20a-5p and XIAP in OS. This result presented that tissue miR-20a-5p expression was low in patients with OS, while tissue XIAP expression was high in them. Through correlation analysis, it was found that LSINCT5 was negatively related to miR-20a-5p, and it was positively correlated with XIAP, and there was a negative relation between the XIAP and miR-20a-5p. This laterally confirmed that LSINCT5 acted as a miR-20a-5p sponge to control XIAP in the development of OS. However, there are still some shortcomings in this study. Firstly, the mechanism of lncRNA-miR-mRNA was studied in this research. Whether LSINCT5 participates in the classical molecular pathway is still unclear. In addition, we have not built the LSINCT5ceRNA network, and we are not clear about other mechanisms of LSINCT5. Finally, the test sample is single in this study. Previous studies have suggested that lncRNA in blood can be used as a potential diagnostic indicator for tumors. Therefore, we hope to carry out bioinformatics analysis in future studies to test the expression of LSINCT5 in the blood of patients with OS to supplement our results.
Conclusion
LncRNA LSINCT5 acts as an oncogene and promotes XIAP-mediated growth and metastasis as a miR-20a-5p sponge in OS.
Acknowledgments
This study was supported by MiR-155 regulates the expression of MMP-8, 9, 13 in spinal tuberculosis, No. 81560359.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yuan T-B, Liu J, Chen S-C, et al. Clinical significance of exosomal long noncoding RNA DANCR as a novel serum-based diagnostic and prognostic biomarker in osteosarcoma. Int J Clin Exp Med. 2019;12(1):423–432.
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Zhang Y, He Z, Li Y, et al. Selection of surgical methods in the treatment of upper tibia osteosarcoma and prognostic analysis. Oncol Res Treat. 2017;40(9):528–532. doi:10.1159/000477251
4. Xu H, Zhu X, Bao H, et al. Genetic and clonal dissection of osteosarcoma progression and lung metastasis. Int J Cancer. 2018;143(5):1134–1142. doi:10.1002/ijc.31389
5. Morrow JJ, Bayles I, Funnell APW, et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat Med. 2018;24(2):176–185. doi:10.1038/nm.4475
6. Simpson S, Dunning MD, de Brot S, Grau-Roma L, Mongan NP, Rutland CS. Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics. Acta Vet Scand. 2017;59(1):71. doi:10.1186/s13028-017-0341-9
7. Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234(11):20721–20727. doi:10.1002/jcp.28678
8. Wei R, Zhang L, Hu W, Wu J, Zhang W. Long non-coding RNA AK038897 aggravates cerebral ischemia/reperfusion injury via acting as a ceRNA for miR-26a-5p to target DAPK1. Exp Neurol. 2019;314:100–110. doi:10.1016/j.expneurol.2019.01.009
9. Yao N, Fu Y, Chen L, et al. Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene. 2019;38(47):7216–7233. doi:10.1038/s41388-019-0904-5
10. Li J, Gao C, Liu C, et al. Four lncRNAs associated with breast cancer prognosis identified by coexpression network analysis. J Cell Physiol. 2019;234(8):14019–14030. doi:10.1002/jcp.28089
11. Zhang Y, Lin S, Yang X, Zhang X. Prognostic and clinicopathological significance of lncRNA MVIH in cancer patients. J Cancer. 2019;10(6):1503–1510. doi:10.7150/jca.28541
12. He W, Lu M, Xiao D. LSINCT5 predicts unfavorable prognosis and exerts oncogenic function in osteosarcoma. Biosci Rep. 2019;39(5):5. doi:10.1042/BSR20190612
13. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
14. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi:10.1093/nar/gkz757
15. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
16. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi:10.1093/nar/gkt1248
17. Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi:10.1093/nar/gkx1067
18. Jiao X, Jiao C, Xu X. Hippo-YAP signaling pathway is associated with the prognosis in children with osteosarcoma. Int J Clin Exp Med. 2019;12(1):589–596.
19. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
20. Long X, Li L, Zhou Q, et al. Long non-coding RNA LSINCT5 promotes ovarian cancer cell proliferation, migration and invasion by disrupting the CXCL12/CXCR4 signalling axis. Oncol Lett. 2018;15(5):7200–7206. doi:10.3892/ol.2018.8241
21. Qi P, Lin WR, Zhang M, et al. E2F1 induces LSINCT5 transcriptional activity and promotes gastric cancer progression by affecting the epithelial-mesenchymal transition. Cancer Manag Res. 2018;10:2563–2571. doi:10.2147/CMAR.S171652
22. Jing L, Lin J, Zhao Y, et al. Long noncoding RNA LSINCT5 is upregulated and promotes the progression of esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(12):5195–5205. doi:10.26355/eurrev_201906_18184
23. Zhang X, Sha M, Yao Y, Da J, Jing D. Increased B-type-natriuretic peptide promotes myocardial cell apoptosis via the B-type-natriuretic peptide/long non-coding RNA LSINCT5/caspase-1/interleukin 1beta signaling pathway. Mol Med Rep. 2015;12(5):6761–6767. doi:10.3892/mmr.2015.4247
24. Tian Y, Zhang N, Chen S, Ma Y, Liu Y. The long non-coding RNA LSINCT5 promotes malignancy in non-small cell lung cancer by stabilizing HMGA2. Cell Cycle. 2018;17(10):1188–1198. doi:10.1080/15384101.2018.1467675
25. Wang LX, Wan C, Dong ZB, Wang BH, Liu HY, Li Y. Integrative analysis of long noncoding RNA (lncRNA), microRNA (miRNA) and mRNA expression and construction of a competing endogenous RNA (ceRNA) network in metastatic melanoma. Med Sci Monit. 2019;25:2896–2907. doi:10.12659/MSM.913881
26. Lv L, Jia J-Q, Chen J. The lncRNA CCAT1 upregulates proliferation and invasion in melanoma cells via suppressing miR-33a. Oncol Res. 2018;26(2):201–208. doi:10.3727/096504017X14920318811749
27. Wu H, Zhou C. Long non-coding RNA UCA1 promotes lung cancer cell proliferation and migration via microRNA-193a/HMGB1 axis. Biochem Biophys Res Commun. 2018;496(2):738–745. doi:10.1016/j.bbrc.2018.01.097
28. Wang L, Luan T, Zhou S, et al. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p. Cancer Med. 2019;8(9):4389–4403. doi:10.1002/cam4.2335
29. Yang R, Fu Y, Zeng Y, et al. Serum miR-20a is a promising biomarker for gastric cancer. Biomed Rep. 2017;6(4):429–434. doi:10.3892/br.2017.862
30. Xu X, Zhu S, Tao Z, Ye S. High circulating miR-18a, miR-20a, and miR-92a expression correlates with poor prognosis in patients with non-small cell lung cancer. Cancer Med. 2018;7(1):21–31. doi:10.1002/cam4.1238
31. Luengo-Gil G, Gonzalez-Billalabeitia E, Perez-Henarejos SA, et al. Angiogenic role of miR-20a in breast cancer. PLoS One. 2018;13(4):e0194638. doi:10.1371/journal.pone.0194638
32. Pu Y, Yi Q, Zhao F, Wang H, Cai W, Cai S. MiR-20a-5p represses multi-drug resistance in osteosarcoma by targeting the KIF26B gene. Cancer Cell Int. 2016;16(1):64. doi:10.1186/s12935-016-0340-3
33. Zhao F, Pu Y, Cui M, Wang H, Cai S. MiR-20a-5p represses the multi-drug resistance of osteosarcoma by targeting the SDC2 gene. Cancer Cell Int. 2017;17(1):100. doi:10.1186/s12935-017-0470-2
34. Ono H, Iizumi Y, Goi W, Sowa Y, Taguchi T, Sakai T. Ribosomal protein S3 regulates XIAP expression independently of the NF-kappaB pathway in breast cancer cells. Oncol Rep. 2017;38(5):3205–3210. doi:10.3892/or.2017.6008
35. Li X, Chen W, Zeng W, Wan C, Duan S, Jiang S. microRNA-137 promotes apoptosis in ovarian cancer cells via the regulation of XIAP. Br J Cancer. 2017;116(1):66–76. doi:10.1038/bjc.2016.379
36. Yang WZ, Zhou H, Yan Y. XIAP underlies apoptosis resistance of renal cell carcinoma cells. Mol Med Rep. 2018;17(1):125–130. doi:10.3892/mmr.2017.7925
37. Hussain AR, Siraj AK, Ahmed M, et al. XIAP over-expression is an independent poor prognostic marker in Middle Eastern breast cancer and can be targeted to induce efficient apoptosis. BMC Cancer. 2017;17(1):640. doi:10.1186/s12885-017-3627-4
38. Liu XG, Xu J, Li F, Li MJ, Hu T. Down-regulation of miR-377 contributes to cisplatin resistance by targeting XIAP in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(5):1249–1257. doi:10.26355/eurrev_201803_14465
39. Zheng J-X, Jiang H, Xiao L-P, Wang -S-S, Li X-D. miR-320 inhibits multidrug resistance of osteosarcoma cells to methotrexate by targeting XIAP. Int J Clin Exp Pathol. 2017;10:368–376.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.