Back to Journals » Cancer Management and Research » Volume 12
LncRNA Lnc712 Promotes Tumorigenesis in Hepatocellular Carcinoma by Targeting miR-142-3p/Bach-1 Axis
Received 21 March 2020
Accepted for publication 4 September 2020
Published 5 November 2020 Volume 2020:12 Pages 11285—11294
DOI https://doi.org/10.2147/CMAR.S254950
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Dan Cui, Caifang Ni
Department of Interventional Radiology, The First Affiliated Hospital of Soochow University, Suzhou City, Jiangsu Province 215006, People’s Republic of China
Correspondence: Caifang Ni
Department of Interventional Radiology, The First Affiliated Hospital of Soochow University, No.188 Shizi Street, Suzhou City, Jiangsu Province 215006, People’s Republic of China
Tel +86-512-65223637
Email [email protected]
Background: It is known that Lnc712 plays an important role in the pathogenesis of breast cancer. However, whether it is involved in hepatocellular carcinoma (HCC) remains unknown. In this study, we aimed to investigate the role and underlying mechanism of Lnc712 in HCC.
Methods: Sixty-four HCC patients were enrolled and followed up for 5 years to analyze the prognostic value of Lnc712 for HCC. HCC cells were transfected with Lnc712 expression vector, Bach-1 expression vector (or siRNA) and miR-142-3p mimic (or inhibitor) to explore the interactions among Lnc712, miR-142-3p and Bach-1. Cell proliferation, migration, invasion and cell cycle were analyzed by CCK-8 assay, transwell assay, wound healing assay and flow cytometry assay, respectively.
Results: The expression of Lnc712 was upregulated in HCC, and the upregulated Lnc712 expression was significantly related to poor overall survival in HCC patients. In HCC cells, Lnc712 interacted with miR-142-3p and upregulated Bach-1, a target of miR-142-3p. In addition, Lnc712 promoted HCC cell proliferation, migration, invasion and cell cycle, while its effects were abolished by miR-142-3p mimic. Moreover, miR-142-3p mimic enhanced HCC cell proliferation, migration, invasion and cell cycle, while its effects were abolished by Bach-1 overexpression. miR-142-3p inhibitor repressed cell proliferation, migration, invasion and cell cycle in HCC cells, while its effects were abolished by Bach-1 knockdown. Furthermore, Lnc712 knockdown remarkably inhibited HCC tumor growth in nude mice.
Conclusion: Lnc712 may promote the development of HCC by targeting the miR-142-3p/Bach-1 axis.
Keywords: hepatocellular carcinoma, Lnc712, Bach-1, miR-142-3p
Introduction
Hepatocellular carcinoma (HCC) as one of the public health challenges is the second most common cause of cancer-related deaths.1 According to the latest GLOBOCAN statistics, HCC in 2018 was responsible for a total of 781,631 deaths (8.2% of all cancer deaths) and affected 841,080 new cases (4.7% of all new cancer cases).2 Despite the advances in cancer diagnosis and treatment, HCC remains one of the most fatal malignancies.3 It has been well established that HBV and HCV infections are the main risk factors for HCC.4–6 In view of the high prevalence of HBV and HCV infections, the incidence of HCC is predicted to further increase in the near future.7 Therefore, novel preventative and treatment approaches are still needed.
Many researchers have been devoted to developing novel anti-cancer drugs from natural products. For example, bitter melon extract has been found to inhibit tumor growth in head, neck and breast cancers.8–10 At the same time, accumulating studies have been also attempted to improve the effectiveness of existing anti-cancer drugs. For instance, methyl-β-cyclodextrin, a cholesterol depleting agent, has been found to enhance the effectiveness of tamoxifen and doxorubicin, two widely used anti-cancer drugs, in melanoma, breast and liver cancers.11,12 Targeted therapy is a novel anti-cancer therapy that aims to suppress cancer development and progression mainly by regulating cancer-related gene expression.13,14 Although existing studies have uncovered a promising prospect of targeted therapies,15 its development closely relies on the identification of effective molecular targets.13,14
Existing studies have revealed that non-coding small RNAs, like LncRNA and microRNA, are implicated in the pathogenesis of cancer. For example, LncRNA MCM3AP-AS1 and KTN1-AS1 can promote the growth of HCC,16,17 while LncRNA MAGI2-AS3 suppresses HCC cell proliferation and migration.18 miR-203 has been found to inhibit tumorigenesis in HCC and breast cancer,19,20 and miR-142-3p represses the proliferation, migration and invasion in HCC and pancreatic cancer cells.21,22 Lnc712 is a recently reported LncRNA which has been identified as a novel oncogenic LncRNA in breast cancer.23 However, whether it is involved in the pathological process of HCC is unknown.
In this study, we aimed to investigate the role and underlying mechanism of Lnc712 in HCC. We found that Lnc712 was upregulated in HCC tissues, and the upregulated expression of Lnc712 predicted poor overall survival in HCC patients. In HCC cells, we further revealed that Lnc712 promoted cell growth, migration and invasion by regulating the miR-142-3p/Bach-1 axis.
Materials and Methods
HCC Patients and a 5-Year Follow-Up
This study was performed according to the 1975 Declaration of Helsinki. Before the admission of patients, this study obtained ethics approval from The First Affiliated Hospital of Soochow University Ethics Committee and was conducted in accordance with the 1975 Declaration of Helsinki. From August 2012 to August 2014, 38 male and 26 female HCC patients were enrolled at the aforementioned hospital.
These 64 patients were all newly diagnosed cases with no initiated therapy by the day of admission. No previous history of malignancy was observed from these patients. These patients were excluded from other severe clinical disorders, such as diabetes, heart disease or severe infections. From the day of admission, all patients were visited monthly for a total of 5 years to monitor their survival. All 64 patients signed written informed consent. The basic characteristics of the HCC patients are listed in Table 1.
 |
Table 1 Basic Characteristics of 64 HCC Patients |
Tissue Collection
Paired HCC and adjacent (within 2 cm around tumors) non-tumor tissues were harvested from each patient. Histopathological examination was performed on all tissue samples to confirm them. Tissues were immediately subjected to RNA isolation after collection.
HCC Cells and Cell Transfections
Human HCC cell line SNU-182 (ATCC, USA) and normal human liver cell line HL-7702 (ATCC) were authenticated by short tandem repeat (STR) profiling using an AmpFLSTR Identifiler PCR Amplification kit (Applied Biosystems, Foster). Cell culture medium was prepared by mixing RPMI 1640 medium (90%), FBS (10%) and 1% penicillin–streptomycin solution. A 5% CO2 incubator was used to cultivate the cells with 95% humidity at 37 °C. Mycoplasma testing was conducted by the LookOut Mycoplasma PCR detection kit (Sigma-Aldrich) once every 2 weeks. Cells used for subsequent experiments were collected at about 80% confluence.
Backbone vectors expressing Lnc712 and Bach-1 were constructed using pcDNA3.1 vector (Sigma-Aldrich). miR-142-3p mimic, inhibitor and negative control (NC) miRNA were obtained from Sangon. Expression vector (1.2 μg) or miRNA (35 nM) was transfected into 108 SNU-182 cells using Lipofectamine 2000 (Invitrogen). Untransfected cells were control (C) cells. NC miRNA- or empty vector-transfected cells were NC cells. At 48 h later, cells were harvested to perform subsequent experiments.
Dual-Luciferase Activity Assay
Backbone luciferase vector of Lnc712 was constructed with pGL3 (Promega Corporation). To explore the interaction between Lnc712 and miR-142-3p, Lnc712 luciferase vector + miR-142-3p mimic (miR-142-3p group) or Lnc712 luciferase vector + NC miRNA (NC group) was transfected into 108 SNU-182 or HL-7702 cells by Lipofectamine 2000. At 48 h later, luciferase activity was measured.
RNA Isolation
SNU-182 (or HL-7702) cells, paired HCC tissues and non-tumor tissues were all subjected to total RNA isolation, which was performed using RNAzol reagent (Invitrogen). To completely remove genomic DNAs, RNA samples were digested with gDNA eraser (Takara) at 37 °C for 2 h.
RT-qPCR
The ExcelRT™ Reverse Transcription Kit (Smobio) was used to reverse transcribe RNA samples into cDNA samples. To perform qPCR, the QuantiFast SYBR Green PCR Kit (QIAGEN) was used to prepare qPCR mixtures with cDNA samples at the template to determine the expression of Lnc712, miR-142-3p and Bach-1 with U6 and GAPDH as internal controls. For example, to measure the expression of miR-142-3p, three steps were performed: 1) adding poly(A) to mature miRNAs; 2) miRNA reverse transcriptions; and 3) qPCR. All steps were completed using the Genecopoeia All-in-One™ miRNA qRT-PCR Reagent Kit. Three replicates were performed for each experiment. Ct values were normalized using the method of 2–ΔΔCq.
The used primers were listed as follows: Lnc712: forward, 5΄-AAATACCTCACCCTCATCTATACCAAC-3΄, reverse, 5΄-TTTCCCGTTGCCATTGAT-3΄; miR-142-3p: forward, 5ʹ-CGCCGTGTAGTGTTTCCTAC-3ʹ, reverse: 5ʹ-CAGTGCAGGGTCCGAGGT-3ʹ; U6, forward: 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ, reverse, 5ʹ-ACGCTTCACGAATTTGCGTGTC-3ʹ; and GAPDH: forward, 5ʹ-CAAGAAGGTGGTGAAGCAGG-3΄, reverse, 5΄-TCAAAGGTGGAGGAGTGGGT-3΄.
Western Blotting
SNU-182 or HL-7702 cells were subjected to the isolation of proteins using RIPA solution (Sangon). Following protein denaturation (95 °C for 10 min), protein separation was performed using SDS-PAGE gel (6%). PVDF membranes were used to transfer proteins and then blocked in PBS (5% non-fat milk) for 2 h at room temperature. Rabbit-derived Bach-1 (1:1000; ab7288, Abcam), c-myc (1:1000; ab32072, Abcam) and GAPDH (1:1000; ab9485, Abcam) primary antibodies were used to incubate the membranes for 16 h at 4 °C, followed by incubation with membranes for 2 h at room temperature with anti-rabbit lgG-HRP (1:1000; ab6721, Abcam) secondary antibody. Signals were produced using ECL (Sangon). Quantity One software (Bio-Rad) was used for data normalization.
Cell Counting Kit-8 (CCK-8)
SNU-182 or HL-7702 cells were subjected to CCK-8 assay using a kit from Dojindo (Japan). A total of 3000 cells in 0.1 mL medium were transferred to each well of a 96-well plate. Cell culture was performed at 37 °C. To monitor cell proliferation, OD values (450 nm) were measured every 24 h for a total of 4 days. At 4 h before the measurement of OD values, CCK-8 solution was added to 10%.
Transwell Assay
Cell migration was assessed by transwell assay. Before the migration assay, the transwell inserts were incubated with 100 μL matrigel (Millipore, USA) for 6 h at 37 °C. Then, 106 SNU-182 or HL-7702 cells were transferred to the upper chamber, and 1 mL culture medium containing 20% FBS was added into the lower chamber. After 48 h of culture, 0.5% crystal violet (Sigma-Aldrich, USA) was used to stain membranes for 20 min. Finally, an optical microscope (Olympus) was used to measure the migration cells.
Wound Healing Assay
A total of 106 SNU-182 or HL-7702 cells were seeded into a 6-well plate. When reaching 90% confluency, the cell monolayers were scratched by a pipette tip to generate the wound. The debris and floating cells were removed through washing with PBS. At 0 and 24 h after scratch, the photographic images were taken from each well.
Cell Cycle Detection
A total of 106 SNU-182 or HL-7702 cells were fixed in 75% ethanol at 4 °C overnight and washed with PBS. Then, cells were stained with FxCycle™ PI/RNase staining solution for 30 min. Finally, the cell cycle was assessed using an Accuri C6 cytometer (BD Biosciences).
In vivo Animal Study
Female nude mice (4–5 weeks old) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd (Shanghai, China). In brief, 5 × 105 HCC cells transfected with Lnc712 siRNA were injected subcutaneously into the flank of nude mice. Six weeks later, all mice were euthanized by injecting 100 mg/kg sodium pentobarbital followed by cervical dislocation, and the tumor size was determined. All animal experiments were conducted at The First Affiliated Hospital of Soochow University Animal Experiment Center and were approved by The First Affiliated Hospital of Soochow University Animal Protection and Use Committee.
Statistical Analysis
All of the above cell culture-related experiments were performed three times. Data were expressed as mean ± SD values of three replicates. Paired tissues were compared by paired t-test. Multiple groups were compared by one-way ANOVA and then Tukey’s test. To perform survival analysis, the 64 patients were divided into two Lnc712 groups (high and low, n = 32) with the median level of Lnc712 in HCC tissues as the cutoff value. Based on follow-up data, survival curves were plotted. A log rank test was used to compare survival curves. p < 0.05 was considered statistically significant.
Results
Overexpression of Lnc712 is Correlated with the Poor Survival of HCC Patients
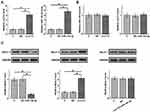
Paired tissues from 64 HCC patients were subjected to RT-qPCR to measure the expression of Lnc712. Compared with non-tumor tissues, HCC tissues exhibited significantly higher expression of Lnc712 (Figure 1A, p < 0.001). Survival analysis showed that, compared with the low Lnc712 expression group, patients in the high Lnc712 expression group exhibited a much higher mortality rate during the 5-year follow-up (Figure 1B).
There is a Direct Interaction Between Lnc712 and miR-142-3p
IntaRNA2.0 was used to predict the interaction between Lnc712 and miR-142-3p. It was observed that they can form strong base pairing (Figure 2A). Dual-luciferase reporter assay analysis showed that, compared with cells transfected with Lnc712 luciferase vector + miR-142-3p mimic (miR-142-3p group), cells transfected with Lnc712 luciferase vector + NC miRNA (NC group) showed significantly lower luciferase activity (Figure 2B, p < 0.05).
Lnc712 Fails to Regulate miR-142-3p Expression but Upregulated Its Target Bach-1 Expression
To explore the interaction between Lnc712 and miR-142-3p, Lnc712 expression vector or miR-142-3p mimic was transfected into SNU-182 cells, and the overexpression of Lnc712 and miR-142-3p was confirmed by RT-qPCR (Figure 3A, p < 0.05). Interestingly, overexpression of Lnc712 or miR-142-3p failed to significantly alter the expression of each other (Figure 3B). Bach-1 is a downstream target of miR-142-3p. To explore the possibility that Lnc712 can sponge miR-142-3p, the role of Lnc712 and miR-142-3p in regulating the expression of Bach-1 was analyzed by western blotting (Figure 3C). Compared with C and NC groups, miR-142-3p overexpression reduced Bach-1 expression, while Lnc712 overexpression increased Bach-1 expression (p < 0.05). Interestingly, co-transfection of Lnc712 expression vector and miR-142-3p mimic failed to significantly affect the expression of Bach-1.
Lnc712 Overexpression Induced Tumorigenesis by Regulating the miR-14-23p/Bach-1 Axis
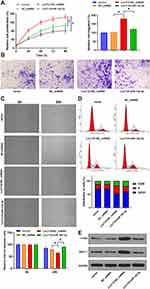
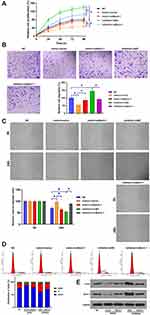
The roles of Lnc712 in tumorigenesis of HCC cells were explored. We found that Lnc712 overexpression promoted proliferation, migration, invasion and cell cycle in SNU-182 cells, while its effects were inhibited by miR-142-3p mimic (Figure 4A–D). Moreover, Lnc712 overexpression upregulated protein levels of c-myc and Bach-1, while miR-142-3p mimic abolished Lnc712-mediated upregulation of these two proteins (Figure 4E). The above results were also confirmed in normal hepatic cell line HL-7702 cells (Figure S1). Moreover, we found that miR-142-3p mimic inhibited proliferation, migration, invasion and cell cycle, and reduced c-myc and Bach-1 proteins in SNU-182 cells, while its effects were repressed by Bach-1 overexpression (Figure 5). miR-142-3p inhibitor enhanced proliferation, migration, invasion and cell cycle, and upregulated c-myc and Bach-1 proteins in SNU-182 cells, while its effects were abolished by Bach-1 knockdown (Figure 5). Collectively, these findings suggest that Lnc712 promotes tumorigenesis in HCC cells by regulating the miR-142-3p/Bach-1 axis.
Lnc712 Knockdown Inhibited HCC Tumor Growth in vivo
The effect of Lnc712 on tumorigenesis in a xenograft study was assessed. Lnc712 siRNA-transfected HCC cells were injected subcutaneously into the flank of the nude mice. The mice were observed over 6 weeks for tumor formation. Results showed that Lnc712 expression was markedly decreased in the xenografts formed by Lnc712 siRNA-transfected cells (Figure 6A). Moreover, the xenograft formed by Lnc712 knockdown had a smaller tumor size compared with that in the control group (Figure 6B and C).
Discussion
The roles of Lnc712, miR-142-3p and Bach-1 in HCC and the possible interactions among them were explored in this study. We found that Lnc712 was upregulated in HCC, and may sponge miR-142-3p to upregulate Bach-1, thereby promoting tumorigenesis in HCC cells (Figure 7).
 |
Figure 7 Schematic diagram of the proposed mechanism. Lnc712 promotes tumorigenesis in HCC cells by regulating the miR-142-3p/Bach-1 axis. |
In a recent study, Cui et al characterized a novel LncRNA named Lnc712 with an oncogenic role in breast cancer.23 It is estimated that Lnc712 may bind heat-shock protein 90 to promote progression of the cell cycle, thereby accelerating cancer cell proliferation.23 Our study is the first to report the upregulation of Lnc712 in HCC. Moreover, an increased proliferation rate of HCC cells was observed after the overexpression of Lnc712. Therefore, Lnc712 may also play an oncogenic role in HCC by promoting cancer cell proliferation, and the development of breast cancer and HCC may share similar molecular pathways.
This study showed that the high expression levels of Lnc712 in HCC tissues were closely correlated with the poor survival of HCC patients. Recent advances in cancer treatment failed to significantly improve the survival of HCC patients.24 As an alternative approach, accurate prediction of patients’ survival may improve treatment outcomes by guiding the selection of therapeutic approaches. Considering that a high level of Lnc712 is correlated with the poor survival in HCC patients, Lnc712 may be a potential diagnostic marker of HCC. However, more clinical trials are needed to confirm the clinical value of Lnc712.
miR-142-3p plays different roles in different types of cancers.25,26 In breast cancer, miR-142-3p targets Bach-1 to suppress cancer metastasis.25 In contrast, miR-142-3p targets FOXO1 to promote cell proliferation and cell cycle progression in prostate cancer.26 In this study, we showed that miR-142-3p targeted Bach-1 to suppress cancer cell proliferation in HCC. Therefore, miR-142-3p may play a role as a tumor suppressor in HCC.
The most interesting finding of this study is that Lnc712 may sponge miR-142-3p to upregulate Bach-1, thereby promoting tumorigenesis in HCC cells. Previous studies have found that Bach-1, as a transcription factor, can promote breast cancer cell migration and invasion by regulating its downstream target genes, such as MMP1, CXCR4, MMP9 and VEGFR.25,27 In addition, the study by Davudian et al found that Bach-1 could regulate the cell cycle pathway by targeting cell cycle-related proteins, such as ITPR2, CALM1 and SQSTM1.28 In this study, we also found that Bach-1 could upregulate the cell cycle regulatory protein c-myc. Taken together, we speculate that Lnc712 and miR-142-3p affect HCC cell growth, migration and invasion by regulating downstream target genes of Bach-1.
Conclusions
In conclusion, the expression of Lnc712 was upregulated in HCC. Lnc712 might sponge miR-142-3p to upregulate Bach-1, thereby promoting cancer proliferation, migration, invasion and cell cycle in HCC cells. Animal experiments are needed to further confirm our findings.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152(4):745–761. doi:10.1053/j.gastro.2016.11.048
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Bertuccio P, Turati F, Carioli G, et al. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67(2):302–309. doi:10.1016/j.jhep.2017.03.011
4. Balogh J, Victor D
5. Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64(1 Suppl):S84–S101. doi:10.1016/j.jhep.2016.02.021
6. Axley P, Ahmed Z, Ravi S, Singal AK. Hepatitis C virus and hepatocellular carcinoma: a narrative review. J Clin Transl Hepatol. 2018;6(1):79–84.
7. Petrick JL, Kelly SP, Altekruse SF, McGlynn KA, Rosenberg PS. Future of hepatocellular carcinoma incidence in the United States forecast through 2030. J Clin Oncol. 2016;34(15):1787–1794. doi:10.1200/JCO.2015.64.7412
8. Bhattacharya S, Muhammad N, Steele R, Kornbluth J, Ray RB. Bitter melon enhances natural killer-mediated toxicity against head and neck cancer cells. Cancer Prev Res (Phila). 2017;10(6):337–344. doi:10.1158/1940-6207.CAPR-17-0046
9. Bhattacharya S, Muhammad N, Steele R, Peng G, Ray RB. Immunomodulatory role of bitter melon extract in inhibition of head and neck squamous cell carcinoma growth. Oncotarget. 2016;7(22):33202–33209. doi:10.18632/oncotarget.8898
10. Muhammad N, Steele R, Isbell TS, Philips N, Ray RB. Bitter melon extract inhibits breast cancer growth in preclinical model by inducing autophagic cell death. Oncotarget. 2017;8(39):66226–66236. doi:10.18632/oncotarget.19887
11. Mohammad N, Malvi P, Meena AS, et al. Cholesterol depletion by methyl-β-cyclodextrin augments tamoxifen induced cell death by enhancing its uptake in melanoma. Mol Cancer. 2014;13(1):204.
12. Mohammad N, Singh SV, Malvi P, et al. Strategy to enhance efficacy of doxorubicin in solid tumor cells by methyl-β-cyclodextrin: involvement of p53 and Fas receptor ligand complex. Sci Rep. 2015;5:11853.
13. Sawyers C. Targeted cancer therapy. Nature. 2004;432(7015):294–297. doi:10.1038/nature03095
14. Wu H-C, Chang D-K, Huang C-T, Shouip H. Targeted therapy for cancer. Journal of Cancer Molecules. 2006;2(2):57–66.
15. Singh S, Chouhan S, Mohammad N, Bhat MK. Resistin causes G1 arrest in colon cancer cells through upregulation of SOCS3. FEBS Lett. 2017;591(10):1371–1382. doi:10.1002/1873-3468.12655
16. Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18(1):28. doi:10.1186/s12943-019-0957-7
17. Zhang L, Wang L, Wang Y, et al. LncRNA KTN1-AS1 promotes tumor growth of hepatocellular carcinoma by targeting miR-23c/ERBB2IP axis. Biomed Pharmacother. 2019;109:1140–1147. doi:10.1016/j.biopha.2018.10.105
18. Yin Z, Ma T, Yan J, et al. LncRNA MAGI2-AS3 inhibits hepatocellular carcinoma cell proliferation and migration by targeting the miR-374b-5p/SMG1 signaling pathway. J Cell Physiol. 2019;234(10):18825–18836. doi:10.1002/jcp.28521
19. Muhammad N, Bhattacharya S, Steele R, Ray RB. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7(36):58595–58605. doi:10.18632/oncotarget.11193
20. Wan D, Shen S, Fu S, et al. miR-203 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC. Anticancer Agents Med Chem. 2016;16(4):414–423. doi:10.2174/1871520615666150716105955
21. Hua S, Liu C, Liu L, Wu D. miR-142-3p inhibits aerobic glycolysis and cell proliferation in hepatocellular carcinoma via targeting LDHA. Biochem Biophys Res Commun. 2018;496(3):947–954. doi:10.1016/j.bbrc.2018.01.112
22. Zhu K, Zhang Z, Zhang H, Wang Z, Wang F. MiR-142-3p targeting NUCKS1 inhibits proliferation and invasion of pancreatic cancer cells. Artif Cells Nanomed Biotechnol. 2020;48(1):415–424. doi:10.1080/21691401.2019.1652629
23. Cui Y, Lu C, Zhang Z, et al. A Long Non-coding RNA Lnc712 Regulates Breast Cancer Cell Proliferation. Int J Biol Sci. 2020;16(1):162–171. doi:10.7150/ijbs.36429
24. Ishaque T, Massie AB, Bowring MG, et al. Liver transplantation and waitlist mortality for HCC and non-HCC candidates following the 2015 HCC exception policy change. Am J Transplant. 2019;19(2):564–572. doi:10.1111/ajt.15144
25. Mansoori B, Mohammadi A, Ghasabi M, et al. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J Cell Physiol. 2019;234(6):9816–9825. doi:10.1002/jcp.27670
26. Tan Y-F, Chen Z-Y, Wang L, Wang M, Liu X-H. MiR-142-3p functions as an oncogene in prostate cancer by targeting FOXO1. J Cancer. 2020;11:1614–1624. doi:10.7150/jca.41888
27. Liang Y, Wu H, Lei R, et al. Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J Biol Chem. 2012;287(40):33533–33544. doi:10.1074/jbc.M112.392332
28. Davudian S, Mansoori B, Shajari N, Mohammadi A, Baradaran B. BACH1, the master regulator gene: a novel candidate target for cancer therapy. Gene. 2016;588(1):30–37. doi:10.1016/j.gene.2016.04.040
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.