Back to Journals » OncoTargets and Therapy » Volume 13
LncRNA LINC01116 Promotes the Development of Colorectal Cancer by Targeting miR-9-5p/STMN1
Authors Bi C, Cui H, Fan H, Li L
Received 2 April 2020
Accepted for publication 7 July 2020
Published 15 October 2020 Volume 2020:13 Pages 10547—10558
DOI https://doi.org/10.2147/OTT.S253532
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Chongyao Bi,1 Hongshuai Cui,2 Haijing Fan,2 Lai Li2
1Department of General Surgery, Jiaozhou Central Hospital of Qingdao, Qingdao 266300, People’s Republic of China; 2Department of General Surgery, The Second Affiliated Hospital of Qingdao University, Qingdao 266041, People’s Republic of China
Correspondence: Lai Li
Department of General Surgery, The Second Affiliated Hospital of Qingdao University, No. 127 Siliunan Road, Shibei District, Qingdao, Shandong Province 266041, People’s Republic of China
Tel +86-532-84961764
Email [email protected]
Purpose: The aim was to research the role of LINC01116 in the prognosis of colorectal cancer (CRC) patients and development of colorectal cancer cells.
Methods: In total 62 colorectal cancer patient tissues and human CRC cell lines (OUMS23, SW116, SW480 and LOVO) were obtained for this study. SiLINC01116, miR-9-5p mimic, LINC01116, oe-STMN1 and their controls were transfected. The qRT-PCR method and Western blot were used to detect the levels of LINC01116, miR-9-5p and STMN1 in tissues and cells. CCK8 assay and flow cytometry were processed for proliferation and apoptosis, respectively. Transwell assay was undertaken to verify invasion and migration. Luciferase assay and pull down assay were processed to confirm the binding relationship among LINC01116, miR-9-5p and STMN1. Immunohistochemistry assay also detected the expression of STMN1. Kaplan–Meier survival curve was used to analyze patient survival rate. Pearson correlation analysis was used to evaluate the regulatory relationship between LINC01116, miR-9-5p and STMN1 in tissues.
Results: LINC01116 was expressed higher in CRC tissues and cells. Patients with higher expression of LINC01116 had worse prognosis. Knockdown of LINC01116 suppressed development of CRC cell. LINC01116 negatively regulated miR-9-5p, while MiR-9-5p was negatively related to STMN1. miR-9-5p mimic could rescue the effect of LINC01116, inhibit migration and invasion, and improve apoptosis of CRC cells. Oe-STMN1 could also rescue the effect of miR-9-5p on the development of colorectal cancer.
Conclusion: LINC01116 promoted the development of colorectal cancer via modulating miR-9-5p/STMN1 axis.
Keywords: colorectal neoplasms, LINC01116, miR-9-5p, STMN1, target
Introduction
Colorectal cancer (CRC) is one of the most frequent malignant tumors worldwide.1 Globally, the mortality and incidence of colorectal cancer ranks third among all genders, which is increasing at a rate of more than 1 million new cases each year.2 Typical symptoms are alternation of diarrhea, constipation, mucus and blood.3 Clinically, comprehensive treatment based on surgery should generally be adopted.4 The treatment measures are decided according to the patient’s general condition and the function of each organ, the location of the tumor, the clinical stage of the tumor, the pathological type, and the biological behavior.5 At present, the treatment of colorectal cancer mainly includes surgery, radiation therapy, chemotherapy, and targeted therapy.6
In recent years, due to the development of molecular biology, the research on colorectal cancer development mechanism is focused on changes of the gene levels. Gene-targeted therapy is a future development trend. Compared with traditional therapy, gene therapy is more targeted and shorter, which can avoid the emergence of drug resistance and systemic toxic reactions.7 Although a large number of genetic mechanisms or protein changes involved in the evolution of colorectal cancer have been discovered and the molecular framework of colorectal cancer evolution has been elucidated, the current research results are far from enough to achieve effective molecular targeted therapy for colorectal cancer clinically.8 Therefore, finding new research strategies and molecular targets is an urgent problem in the field of basic tumor research and clinical treatment.
LncRNA is a non-coding RNA closely related to tumor genesis process, attracting investigator’s attention. LINC01116 is more highly expressed in many tumors, which promotes proliferation and invasion of cells.9 For instance, lncRNA-LINC01116 could target Vascular Endothelial Growth Factor A (VEGFA), and then effect the cell proliferation, invasion, migration, cycle, and angiogenesis of brain glioma tumor.10 In addition, Zhang et al11 confirmed that LINC01116 improves development of osteosarcoma cancer by regulating the expression of miR-520a-3p and Interleukin 6 Receptor (IL6R) and participating in the Jak-stat signaling pathway.11 Hu et al12 also referred that LINC01116 overexpression was a biomarker of poor prognosis for breast cancer patients. Moreover, LINC01116 was higher expressed in head and neck squamous cell carcinoma, which could promote cell invasion of this disease by involving the epithelial mesenchymal transition pathway.13 Thereby, LINC01116 is important for development of various cancers. However, the role and mechanism of LINC01116 in CRC has not been reported. The aim of this research was to investigate the expression of LINC01116 in colorectal cancer, its effect on patient prognosis, and the molecular mechanism of LINC01116 in CRC cells. Interestingly, based on the bioinformatics online prediction software (Starbase and targetscan), LINC01116 was closely related to miR-9-5p/STMN1 expression. MiR-9-5p has been verified to participate in various cancers, such as non-small-cell lung cancer,14 thyroid cancer,15 and prostate cancer.16 Besides, STMN1 protein is a highly conserved intracellular protein. During the mitotic interval, the active state of STMN1 protein promotes microtubule depolymerization.17 The expression level of STMN1 in tumors is directly related to the prognosis of patients, and the expression of stathmin protein in tumor cells affects the sensitivity of vinblastine and paclitaxel drugs.18
Based on the above evidence, LINC01116 plays a critical role in tumor development. However, the detail of the molecular mechanism of LINC01116 in CRC is still unclear. Thereby, exploring the role of LINC01116 and related factors may lay the foundation for CRC treatment. In this study, the expression of LINC01116 in CRC tissues and cells was detected, and the relationship between LINC01116 level and patient survival rate was analyzed. Moreover, the effect of LINC01116 expression on migration, invasion, and apoptosis was also researched. Finally, the regulation of LINC01116, miR-9-5p and STMN1 was explored, and these factors in CRC development were also confirmed.
Materials and Methods
Clinical Samples
In total 62 patients who were diagnosed with colorectal cancer and treated with surgery from October 2018 to June 2019 in our hospital were included. This study was approved by the ethics committee of The Second Affiliated Hospital of Qingdao University, and was conducted in accordance with the Declaration of Helsinki. All patients’ families were informed of the study and signed informed consent. CRC tissues and normal adjacent tissues (Less than 3 cm adjacent to cancer) were obtained, confirmed by biopsy and stored immediately at −80°C in a refrigerator. Clinical characteristics including ages, gender, tumor size, TMN stage, local invasion, lymphatic metastasis and overall survival were recorded and analyzed. Based on the median expression of LINC01116, all patients were divided into two groups. Ages, gender, local invasion and lymphatic metastasis between the two groups were not significantly different (Table 1). However, patients with LINC01116 high expression always had larger tumor size and more serious TMN stage.
 |
Table 1 The Correlation Between LINC01116 Expression and COAD Clinical Pathology |
Cell Culture and Transfection
CRC cell lines [OUMS23 (JCRB1022, Epithelial-like, chondrosarcoma), SW116 (ATCC® CCL-228, Colon adenocarcinoma at stage III), SW480 (ATCC® CCL-228, Dukes’B type, colon adenocarcinoma, established from primary colon adenocarcinoma) and LOVO (ATCC® CCL-229, Dukes’ C type, stage IV, colorectal adenocarcinoma)] and human colon epithelial cell line (NCM460, Epithelial-like) were purchased from Beina Chuanglian Biotechnology Research Institute (Beijing, People's Republic of China). Based on these cell lines, the expression and mechanism of LncRNA LINC01116 in various colon cancer cell lines can be more completely presented and verified. The cell lines were cultured in DMEM medium with 10% FBS in a cell incubator (37°C, 5% CO2, Thermo Fisher Scientific, USA). The sequences of siLINC01116-1, siLINC01116-2, siLINC01116-3 and siCtrl were synthesized by Shanghai Biotech Bioengineering Co., Ltd. LOVO and SW116 cells in logarithmic growth phase were seeded in 6-well plates with 2 × 105 cells per well. All cells were randomly grouped into the siLINC01116 group and the negative control group, with three replicates in each group. For each group, cells were routinely incubated for 24 hours before adding transfectants. All operations were performed strictly according to the instructions of Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). The cells of each group were cultured in a cell incubator at 37°C, 5% CO2, and saturated humidity. Similarly, miR-9-5p mimic, LINC01116, oe-STMN1 and their controls were also transfected as above steps.
qRT-PCR
The level of LINC01116 in tissues and cells was detected by qRT-PCR. T TRIzol reagent was added to tissues or cells. After sufficient lysis, chloroform was added, and the samples were centrifuged at 12,000 r/min for 5 minutes on ice. The supernatant was taken and added with isopropanol. After precipitation, the precipitate was washed with 75% ethanol and the RNA was collected, and an appropriate amount of DEPC water was added to dissolve it. The NanoDrop 2000 ultra-micro spectrophotometer was used to detect the extracted total RNA concentration. The extracted total RNA was reverse transcript into cDNA using RR047A PrimeScriptTM RT reagent Kit (TaKaRa). GADPH was used as an internal reference. RR420A SYBR Premix Ex Taq (TaKaRa) was used to perform qRT-PCR, and the relative expression of lncRNA LINC01116 was calculated using the 2-ΔΔCt method. GADPH was the reference for analysis of qRT-PCR results. The primer sequences were shown in Table 2. The primers sequences of lncRNA LINC01116 were as follows: (up-stream) 5ʹ-CTTCTTTCCCTCCAAGTGAT-3ʹ, (down-stream) 5ʹ-TTAGCAAGTCAGCAAGTCCT-3ʹ. Primer 5.0 was used for primer sequence design. Both design and synthesis processes of all primers were completed by GenScript Biotechnology Co., Ltd. (People's Republic of China).
 |
Table 2 Primer Sequences for qRT-PCR |
CCK8 Assay for Proliferation
The CRC cells were inoculated into a 96-well plate with cell suspension (100 μL/well). Transfection was performed at a density of 50%. After 48 hours of transfection, the culture medium was removed from each well of the 96-well plate. Then, 10 μL of Cell Counting Kit-8 (CCK8) solution (Beyotime Biotechnology, People's Republic of China) was supplied to each well, and the culture plate was then incubated for 2 hours. After termination of the culture, the absorbance was measured at 450 nm with a Microplate reader (Thermo Fisher Scientific, Shanghai, People's Republic of China) to determine cell viability.
Flow Cytometry for Apoptosis
Two groups of single cell suspensions were prepared and adjusted to a cell concentration of 106/mL. Then, 0.5 mL of cell suspension was taken, centrifuged at 1000 r/min for 4 minutes, and precipitation was obtained. Next, 0.5 mL of Binding Buffer was used to resuspend the cells, and 1 μL of fluorescently labeled annexin V reagent was added. After mixing, the samples were continually kept for 20 minutes in the dark at room temperature before 5 μL of PI reagent was added and cultured at 4° C for 5 minutes avoiding the light. Finally, 0.5 mL of Binding Buffer was added to the sample and analyzed by flow cytometry.
Transwell Assay for Migration and Invasion
Matrigel was melted, diluted, added to the Transwell upper chamber (Corning, USA), and equilibrated at 37° C for 5 hours. However, matrigel was not used for the cell migration experiment. In total 100 μL of fetal bovine serum-free cell suspension (5.0 × 105 cells/mL) was aspirated and added to the upper chamber, while 600 μL of medium containing 10% fetal bovine serum was added to the lower chamber. After culturing for 24 hours, the remaining liquid was discarded and matrigel and upper chamber cells were removed with a swab. Samples in the submembranous compartment were fixed by paraformaldehyde for 30 minutes, air-dried, and stained with 0.1% crystal violet. Cells of five regions were selected, photographed and calculated under an inverted microscope (100 ×, Olympus, Japan), and the average value of cell numbers was taken.
Luciferase Assay
Fragments containing the wild-type 3’ end noncoding region (3ʹUTR) or mutant 3ʹUTR of the LINC01116 were constructed. Enzyme cutting sites were designed at both ends, and a luciferase reporter gene plasmid (pGL3-promoter, Promega, USA) was inserted to construct pGL3-3ʹUTR wt, pGL3-3ʹUTR mut. HEK293 cells with good growth status were taken. After plasmid transfection for 48 hours, samples were lysed, luciferase substrate was added, and luciferase activity was detected according to instructions of the manufacturer.
Pull-Down Assays
Based on the transcript sequence of lncRNA LINC01116, primers were designed at the 5ʹ and 3ʹ ends, respectively. In order to obtain the lncRNAGAS5 transcript for the positive and negative strand templates, a T7 promoter sequence was added to the 5ʹ end of the primer. Using pcDNA3.1 LINC01116 plasmid DNA as a template, DNA fragments were amplified using positive strand template primers and negative strand templates, respectively. Then, transcription products were randomly labeled with biotin and biotin RNA transcribed in vitro was purified. RNA pull-down was undertaken as reported previously.19 The pull-down products were detected by Western blotting for specific antibodies. The experiment was also undertaken to confirm the binding between miR-9-5p and STMN1.
Western Blot Assay
The cells of each group were collected, and the total protein was extracted after routine lysis, and the adjusted protein concentration was consistent. Then, 40 μg of total protein was aspirated, transferred to PVDF after SDS-PAGE electrophoresis, and blocked with 5% skimmed milk powder. After incubation with the primary antibody (Cellular Signaling Technology, USA), the secondary antibody (Beijing Zhongshan Jinqiao Biotechnology Co., Ltd., People's Republic of China) was added the next day, and the sample was cultured at 37°C for 2 hours. The ECL working solution was added to the sample, the gel imager was used to photo, and the gray value was analyzed by Quantity One. GAPDH was the internal reference.
Immunohistochemistry Assay
All specimens were fixed with formalin at room temperature, dehydrated with alcohol, xylene transparentized, embedded with paraffin immersion wax, and sectioned with paraffin microtome. The samples were placed in an oven at 65° C. Sections were dewaxed with conventional xylene and dehydrated with gradient alcohol. After the antigen was repaired, the sections were naturally cooled in the buffer for 60 minutes before 3% H2O2 was supplied dropwise to the sections and incubated, and left at 37° C for 10 minutes to block endogenous peroxidase. After the serum was blocked, a primary antibody (1: 100) was added dropwise to the sample and placed in a wet box at 4° refrigerator for a whole night. The biotinylated secondary antibody was supplied dropwise and cultured at 37°C for 20 minutes. After the addition of horseradish-labeled streptavidin working solution, DAB coloration and hematoxylin counterstaining were performed. After the sample was dehydrated and transparent, the excess xylene was wiped off. Finally, the sample was sealed, observed and photographed.
Statistical Analysis
The software package SPSS 20.0 was used to analyze the data. Measurement data are shown as M ± SD (xˉ± s). Comparisons between groups were undertaken by t-test. Kaplan-Meier survival curve is used to analyze patient survival rate and survival time. Pearson correlation analysis was used to evaluate the regulatory expression among LINC01116, miR-9-5p and STMN1 in tissues. P<0.05 was the threshold of statistically significant difference.
Results
LINC01116 is Highly Expressed in Colorectal Cancer Tissues and Cells
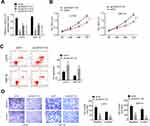
The expression of LINC01116 in CRC tissues and cells was detected by qRT-PCR. It is worth noting that LINC01116 had higher levels in tumor tissues than normal (P<0.0001, Figure 1A). Moreover, LINC01116 had higher levels in tumor tissues at stage I–II than that of patients at stage III–IV (P<0. 01, Figure 1B). Survival curve analysis indicated that patients with highly expressed LINC01116 have poor prognosis (Figure 1C). Similarly results were also obtained in vitro. LINC01116 expression was higher in human CRC cell lines (OUMS23, SW116, SW480 and LOVO) than that in human normal colon epithelial cell line (NCM460) (Figure 1D). It was worth noting that LINC01116 is expressed highest in SW116 and LOVO cell lines. Thereby, the two cell lines were used for further experiments.
LINC01116 Knockdown Inhibited Development of CRC Cells
Knockdown of LINC01116 was performed in LOVO, SW116 cell lines, and the knockdown efficiency was detected. All three knock-down sequences could reduce the expression of LINC01116, indicating that the transfections were successful (Figure 2A). According to the transfection efficiency, siLINC01116-1 was selected for subsequent experiments. After transfection, the proliferation was significantly decreased (Figure 2B), while cell apoptosis was improved in both LOVO and SW116 cell lines (Figure 2C). In addition, migration, and invasion of CRC cells were also suppressed by siLINC01116 (Figure 2D). Thereby, we inferred that LINC01116 knockdown inhibited development of CRC cells.
LINC01116 Targeted miR-9-5p
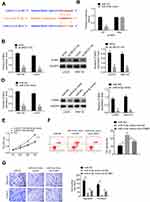
Based on the bioinformatics online prediction software (Starbase and targetscan), the binding site of LINC01116 and miR-9-5p is presented in Figure 3A. Binding sites were detected by a double luciferase reporter assay. After miR-9-5p is overexpressed, the luciferase activity of wild-type lncRNA LINC01116 is significantly inhibited, but the luciferase activity of mutant IncRNA LINC01116 is not significantly affected (Figure 3B). The data show that miR-9-5p can directly bind to lncRNA LINC01116 binding site and affect luciferase activity. To determine the possible direct interaction between LINC01116 and miR-9-5p, a T7-LINC01116 nucleic acid probe was designed and constructed for RNA pull-down experiments in this study. It was found that LINC01116 could significantly pull-down miR-9-5p (Figure 3C). siLINC01116 decreased the expression of miR-9-5p in both SW116 and LOVO cell lines (Figure 3D). Rescue assay was processed to verify the effect of LINC01116 and miR-9-5p on colorectal cancer cells. Based on the transfection, cells were divided into Ctrl, LINC01116+miR-NC and LINC01116+miR-9-5p mimic groups. As the results show, LINC01116 increased the proliferation, migration, and invasion, but inhibited cell apoptosis. Additionally, miR-9-5p rescued the effect of LINC01116 on the above progress (Figure 3E-G).
STMN1 Was the Target of miR-9-5p
Starbase has an online prediction database for the binding site of STMN1 and miR-9-5p, and the binding site is shown in Figure 4A. The result of luciferase assay showed that miR-9-5p could bind to STMN1 binding site and affect luciferase activity (Figure 4B). QRT-PCR and Western blot was processed to detect the STMN1 level after siLINC01116 or miR-9-5p mimic transfection. In both LOVO and SW116 cell lines, siLINC01116 and miR-9-5p could suppress the expression of STMN1 (Figure 4C and D). The rescue assay also verified that up-regulated miR-9-5p could promote apoptosis and inhibit proliferation, migration and invasion of CRC cells, while oe-STMN1 rescued the function of miR-9-5p mimic (Figure 4E-G).
Correlation Between LINC01116 and miR-9-5p/STMN1 Expression in Colorectal Cancer Tissues
miR-9-5p and STMN1 expression was detected by qRT-PCR in CRC tissues and normal tissues. As represented in Figure 5A, miR-9-5p was significantly lower expressed in CRC tissues. However, STMN1 was expressed higher in tumor tissues (Figure 5B). The results of IHC assay were similarto that of qRT-PCR (Figure 5C). Moreover, Pearson correlation analysis was used to evaluate the expression relationship between LINC01116, miR-9-5p and STMN1 in tissues. LINC01116 negatively modulated the miR-9-5p level (Figure 5D). LINC01116 was negatively related to STMN1, while MiR-9-5p was negatively related to STMN1 expression (Figure 5E and F).
Discussion
LINC01116 is mainly used in various cancers for early diagnosis, targeted therapy, and observation of prognosis. In this study, LINC01116 was highly expressed in CRC tissues and cells. Patients with higher LINC01116 level always had poor prognosis. Knockdown of LINC01116 inhibited proliferation, migration, and invasion of CRC cells. LINC01116 negatively regulated miR-9-5p, while MiR-9-5p was negatively related to STMN1 expression.
LINC01116 (GC id: GC02M176629) has been reported to regulate numbers of miRNAs and genes in various cancers. In breast cancer, it could directly combine with miR-145, then improve the expression of ESR1 (GC id: GC06P151656), and promote the development of breast cancer.20 Interestingly, Paun et al21 analyzed a comprehensive profile of epigenetic changes in colorectal cancer and concluded that methylation of ESR1 was the biomarker of colorectal cancer.Furthermore, LINC01116 could regulate the expression of miR-520a-3p/IL6R axis, and then improve migration and proliferation of osteosarcoma cells.22 It worth noting that miR-520a-3p has been confirmed to inhibit invasion, promote apoptosis and decelerate tumor growth by regulating the expression of EGFR (GC id: GC07P055019).23 Moreover, down-regulated IL6R (GC id: GC01P154405) could inhibit migration and invasion by interfering with the expression of IL-6.24 Interestingly, LINC01116 was confirmed to contribute to gefitinib resistance in non-small cell lung cancer through regulating IFI44.25 Thereby, this LncRNA may have a role in drug resistance as well. In this study, CRC patients with higher LINC01116 levels had poor prognosis. Knockdown of LINC01116 was confirmed to inhibit development of CRC cells. Based on the above evidence, we speculated that LINC01116 played a critical role in CRC development.
In addition, lncRNA LINC01116 was confirmed to target miR-9-5p (GC id: GC01M156420) in this study. It has been found that miR-9-5p may play a protocarcinogenic or tumor suppressive effect in tumors, and its functions have not been fully elucidated. As reported in previous studies, miR-9-5p targeted StarD13 (GC id: GC13M033103), and then improved the development of prostate cancer.26 Similarly, miR-9-5p was also more highly expressed in hepatocellular carcinoma, which could regulate SOX11 (GC id: GC02P005704) expression and then improve hepatocellular carcinoma growth.9 However, low expressed miR-9-5p was also verified and researched in different cancers. For instance, Zhang et al27 verified that miR-9-5p inhibited the expression of FOXP2 (GC id: GC07P114086) and further suppressed proliferation of glioma cells. Also, forced expression of miR-9-5p could protect liver from ischemia reperfusion-induced hepatic injury.28 More importantly, Wang et al29 found that up-regulated miR-9-5p inhibit proliferation and improve apoptosis in colorectal cancer by targeting PAK4 (GC id: GC19P039125). Moreover, miR-9-5p was confirmed to regulate some other targets, such as POU2F,30 FOX4, TGFBR2,31 and BCL2L11 in previous studies. Thereinto, POU2F1 could promote growth and metastasis of hepatocellular carcinoma through the FAT1 signaling pathway.32 FAT1 signaling pathway was also a critical pathway in colorectal cancer development.33 Moreover, Ogino et al34 confirmed that TGFBR2 mutation was closely related with CpG island methylator phenotype in microsatellite instability-high colorectal cancer. Importantly, BCL2L11 was regarded as an apoptosis-associated gene.35 Our findings are consistent with previous studies. miR-9-5p mimic could rescue the effect of LINC01116, inhibit migration and invasion, and improve apoptosis of colorectal cancer cells. Thereby, LINC01116 improves the progression of colorectal cancer by inhibiting the expression of miR-9-5p.
Furthermore, STMN1 (GC id: GC01M025884) was the target of miR-9-5p in this study. Interestingly, oe-STMN1 could rescue the effect of miR-9-5p on the development of colorectal cancer cells. In previous studies, STMN1 has been verified to promote biological behavior and invasion in various cancers, such as endometrial carcinoma,36 cutaneous squamous cell carcinoma37 and gastric cancer.38 Moreover, high expression of STMN1 was also regarded as a predictor of progression and poor survival of colorectal cancer patients.39 In 2009, Ogino et al40 processed a cohort study and found that the expression of STMN1 was closely related to survival time of colorectal cancer patients. Tan et al41 undertook a proteomic analysis, and revealed that STMN1 was more highly expressed in both colorectal tumors and metastatic tissues. Additionally, overexpressed STMN1 improved invasion and proliferation of colorectal cancer cells and also decreased overall survival of patients.42 It was worth mentioning that knockdown of STMN1 could inhibit cell adherence and metastasis and also improve chemoresponse to therapeutic medicine, such as 5-Fu.43 More importantly, our results in vivo also confirmed the correlation between LINC01116 and miR-9-5p/STMN1 expression in colorectal cancer.
Furthermore, LINC01116 was found to be involved in different pathways, including the hippo signaling pathway and Jak-stat signaling pathway.44,45 It is worth noting that inactivating the hippo signaling pathway could promote stemness and chemotherapeutic resistance of colorectal cancer.46 Moreover, it could also promote proliferation and tumorigenicity of colorectal cancer.47 Besides, the JAK-STAT signaling pathway was closely related with proliferation and apoptosis of colorectal cancer.48 Thereby, LINC01116 was important for colorectal cancer development. However, there were limitations in this study. LINC01116 was related with different signaling pathways in this disease. In addition, the related pathways were complex and interacted with each other. However, a figure as a systematic view needed more research foundation, which will be the focus of our further research.
In conclusion, LINC01116 promoted the development of colorectal cancer via regulating miR-9-5p/STMN1 expression. It could be applied for treatment of colorectal cancer as a novel target.
Highlights
- LINC01116 was expressed more highly in colorectal cancer cells and tissues.
- Patients with more highly expressed LINC01116 always showed poor prognosis.
- Knockdown of LINC01116 suppressed progression of colorectal cancer cells.
- LINC01116 was negatively regulated by miR-9-5p.
- MiR-9-5p was negatively related to STMN1 expression.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pang X, Li R, Shi D, et al. Knockdown of Rhotekin 2 expression suppresses proliferation and induces apoptosis in colon cancer cells. Oncol Lett.
2. Wilkins T, Mcmechan D, Talukder A. Colorectal cancer screening and prevention. Am Fam Physician. 2018;97:658–665.
3. Gigola G, Zwenger A, Maturi HV, Perdigón G. 155P effect of probiotics on survival in a rat colorectal cancer model treated with capecitabine. Ann Oncol. 2014;25:iv54. doi:10.1093/annonc/mdu325.5
4. Xu J, Qin J, Wang S, et al. Chinese guidelines for the diagnosis and comprehensive treatment of hepatic metastasis of colorectal cancer. J Cancer Res Clin Oncol. 2011. 137:1379–1396.
5. Gu J. The necessary perseverance of surgery for the treatment of locally advanced colorectal cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2018;21(241):2018.
6. Daniela R, Adhemar L-F, F. MS. Predictive biomarkers in colorectal cancer: from the single therapeutic target to a plethora of options. Biomed Res Int. 2016;1–12.
7. Wang CH, Ranganath SH. Editorial [Hot topic: current formulations and techniques of drug/gene delivery for targeted therapy and tissue Engineering (Executive Editors: chi-Hwa Wang and Sudhir H. Ranganath)]. Curr Pharm Des. 2010;16(21):2296–2297. doi:10.2174/138161210791920513
8. Waldner MJ, Neurath MF. The molecular therapy of colorectal cancer. Mol Aspects Med. 31:0–178.
9. !!! INVALID CITATION !!!, !!! INVALID CITATION !!!
10. Ye J, Zhu J, Chen H, et al. A novel lncRNAmmINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int J Cancer. 2020;2020.
11. Zhang B, Yu L, Han N, et al. Corrigendum to “LINC01116 targets miR-520a-3p and affects IL6R to promote the proliferation and migration of osteosarcoma cells through the Jak-stat signaling pathway. Biomed Pharmacother. 2018;107:2018.
12. Hu H-B, Chen Q, Ding S-Q. LncRNA LINC01116 competes with miR-145 for the regulation of ESR1 expression in breast cancer. Eur Rev Med Pharmacol Sci. 2018;2018.
13. Wu J, Chen Z, Zhang L, Cao J, He D. Knockdown of LINC01116 inhibits cell migration and invasion in head and neck squamous cell carcinoma through epithelial-mesenchymal transition pathway. J Cell Biochem. 2019;2019.
14. Yang Y, Kai C, Zhou Y, Hu Z, Shuai C, Huang Y. Application of serum microRNA-9-5p, 21-5p, and 223-3p combined with tumor markers in the diagnosis of non-small-cell lung cancer in Yunnan in southwestern China. Onco Targets Ther. 2018;11:587–597.
15. Chen W, Yu J, Xie R, Zhong M. miR-9-5p increases sensitivity to cisplatin in thyroid cancer cells by down-regulating BRAF expression. J Biomater Tissue Eng. 2019;2019.
16. Chen L, Hu W, Li G, Guo Y, Wan Z, Yu J. Inhibition of miR-9-5p suppresses prostate cancer progress by targeting StarD13. Cell Mol Biol Lett. 2019;24(1):2019. doi:10.1186/s11658-019-0145-1
17. Aronova A, Min IM, Crowley MJP, et al. STMN1 is overexpressed in adrenocortical carcinoma and promotes a more aggressive phenotype in vitro. Ann Surg Oncol. 2017;2017.
18. Wang S, Akhtar J, Wang Z. Anti-STMN1 therapy improves sensitivity to antimicrotubule drugs in esophageal squamous cell carcinoma. Tumour Biol J Int Soc Oncodevel Biol Med. 2015;36:7797–7806.
19. Jiang N, Wang X, Xie Y, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. S0304383517303907.
20. Hu HB, Chen Q, Ding SQ. LncRNA LINC01116 competes with miR-145 for the regulation of ESR1 expression in breast cancer. 2018;22(1987):2018.
21. Paun BC, Mori Y, Sato F, et al. Epigenomic profiling of human colon adenomas. 2006;66:51.
22. Butian Z, Lili Y, Ning H, Zhenzhen H, Shuang W. LINC01116 targets Mir-520a-3p and affectsIL6R to promote the proliferation and migration of osteosarcomacells through the Jak-Stat signaling pathway.
23. Zhang R, Liu R, Liu C, Niu Y, Chen X. A novel role for MiR-520a-3p in regulating EGFR expression in colorectal cancer. Cell Physiol Biochem. 2017;42;1559–1574. doi:10.1159/000479397
24. Rokavec M, Öner MG, Li H, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;2014.
25. Wang H, Lu B, Ren S, et al. Long Noncoding RNA LINC01116 contributes to gefitinib resistance in non-small cell lung cancer through regulating IFI44. Mol Ther Nucleic Acids. 2020;19(218–227):2020. doi:10.1016/j.omtn.2019.10.039
26. Chen L, Hu W, Li G, Guo Y, Wan Z, Yu J. Inhibition of miR-9-5p suppresses prostate cancer progress by targeting StarD13. Cell Mol Biol Lett. 24.
27. Zhang H, Li Y, Tan Y, et al. MiR-9-5p inhibits glioblastoma cells proliferation through directly targeting FOXP2 (Forkhead Box P2). Front Oncol. 2019;9:1176. doi:10.3389/fonc.2019.01176
28. Liao X, Zhou S, Zong J, Wang Z. Sevoflurane exerts protective effects on liver ischemia/reperfusion injury by regulating NFKB3 expression via miR-9-5p. Exp Ther Med. 2019;17:2632–2640. doi:10.3892/etm.2019.7348
29. Wang M, Gao Q, Chen Y, Li Z, Yue L, Cao Y. PAK4, a target of miR-9-5p, promotes cell proliferation and inhibits apoptosis in colorectal cancer. Cell Mol Biol Lett. 2019;24:58. doi:10.1186/s11658-019-0182-9
30. Xie CH, Cao YM, Huang Y, et al. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F1 expression. Tumor Biol. 2016;2016.
31. Fierro-Fernandez M, Busnadiego O, Sandoval P, et al. miR-9-5p suppresses pro-fibrogenic transformation of fibroblasts and prevents organ fibrosis by targeting NOX4 and TGFBR2. EMBO Rep. 2015;16:1358–1377. doi:10.15252/embr.201540750
32. Hong YZ, Guan YC, Shi PW, Yu C, Jian PH. POU2F1 promotes growth and metastasis of hepatocellular carcinoma through the FAT1 signaling pathway. Am J Cancer Res. 2017;7:1665–1679.
33. Karim MA, Shan LQ, Ze-Qun LI, et al. Molecular mechanism of αvβ6-ERK signaling pathway negatively regulates Fat-1 expression and promotes colon cancer invasion. Chin J Curr Advan General Surg. 2016;2016.
34. Ogino S, Kawasaki T, Ogawa A, Kirkner GJ, Loda M, Fuchs CS. TGFBR2 mutation is correlated with CpG island methylator phenotype in microsatellite instability-high colorectal cancer. Hum Pathol. 2007;38:614–620.
35. Kopke S, Buhrke T, Lampen A. miRNA expression in human intestinal Caco‐2 cells is comparably regulated by cis‐ and trans‐fatty acids. Lipids. 2015;50:227–229.
36. He X, Liao Y, Lu W, Xu G, Wan X. Elevated STMN1 promotes tumor growth and invasion in endometrial carcinoma. Tumor Biol. 2016;37:9951–9958. doi:10.1007/s13277-016-4869-5
37. Li X, Wang L, Li T, You B, Cao X. STMN1 overexpression correlates with biological behavior in human cutaneous squamous cell carcinoma. Pathol Res Pract. 2015;211:816–823. doi:10.1016/j.prp.2015.07.009
38. Bai T, Yokobori T, Altan B, Ide M, Kuwano H. High STMN1 level is associated with chemo-resistance and poor prognosis in gastric cancer patients. Br J Cancer. 2017;116:1177–1185. doi:10.1038/bjc.2017.76
39. Wei WU. Role of Stathmin-1 in colorectal cancer metastasis and chemo-resistance. 2014;2014.
40. Ogino S, Nosho K, Baba Y, et al. A cohort study of STMN1 expression in colorectal cancer: body mass index and prognosis. Am J Gastroenterol. 2009;104(8):2047–2056. doi:10.1038/ajg.2009.281
41. Tan HT, Wu W, Ng YZ, et al. Proteomic analysis of colorectal cancer metastasis: stathmin-1 revealed as a player in cancer cell migration and prognostic marker. J Proteome Res. 2012;11(2):1433–1445. doi:10.1021/pr2010956
42. Guo F, Luo Y, Mu YF, Qin SL, Zhong M. miR-193b directly targets STMN1 and inhibits the malignant phenotype in colorectal cancer. Am J Cancer Res. 2016;6:2463–2475.
43. Wu W, Tan XF, Tan HT, Lim TK, Chung MCM. Unbiased proteomic and transcript analyses reveal that Stathmin-1 silencing inhibits colorectal cancer metastasis and sensitizes to 5-fluorouracil treatment. Mol Cancer Res. 2014;12(12):1717–1728. doi:10.1158/1541-7786.MCR-14-0088-T
44. Liang Y, Ma YY, Li LL, et al. Effect of long non-coding RNA LINC01116 on biological behaviors of non-small cell lung cancer cells via the hippo signaling pathway. J Cell Biochem. 2018;2018.
45. Butian Z, Lili Y, Ning H, Zhenzhen H. LINC01116 targets miR-520a-3p and affects IL6R to promote the proliferation and migration of osteosarcoma cells through the Jak-stat signaling pathway. Biomedecine Pharmacotherapie. 2018;2018.
46. Wang X, Sun D, Tai J, et al. TFAP2C promotes stemness and chemotherapeutic resistance in colorectal cancer via inactivating hippo signaling pathway. J Exp Clin Cancer Res. 2018;37:27. doi:10.1186/s13046-018-0683-9
47. Xu W, Di S, Jiandong T, Si C, Sen H. ZNF280A promotes proliferation and tumorigenicity via inactivating the hippo-signaling pathway in colorectal cancer. Mol Ther Oncol. 2019;2019.
48. Li L-X, Lam I-H, Liang -F-F, Yi S-P, Ye L-F. MiR-198 affects the proliferation and apoptosis of colorectal cancer through regulation of ADAM28/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci. 2019;2019.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.