Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 14
LncRNA LINC00673 is Downregulated in Diabetic Retinopathy and Regulates the Apoptosis of Retinal Pigment Epithelial Cells via Negatively Regulating p53
Received 23 December 2020
Accepted for publication 10 August 2021
Published 13 October 2021 Volume 2021:14 Pages 4233—4240
DOI https://doi.org/10.2147/DMSO.S298185
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ming-Hui Zou
Yan Cheng, Yanxia Zhu, Linli Ma
Department of Ophthalmology, The Second People’s Hospital of Lianyungang, Lianyungang 222000, Jiangsu Province, People’s Republic of China
Correspondence: Linli Ma
Department of Ophthalmology, The Second People’s Hospital of Lianyungang, Lianyungang 222000, Jiangsu Province, People’s Republic of China
Email [email protected]
Background: Long noncoding RNA (LncRNA) LINC00673 has been proven to play critical roles in cancer biology, while its role in other diseases is unknown. It has been reported that LINC00673 could interact with p53, a critical player in diabetes and diabetic complications, suggesting that LINC00673 may also participate in diabetic retinopathy (DR). This study aimed to investigate the role of LINC00673 in DR.
Methods: The present study included 3 groups of participants, including DR group, diabetes (DB) group, and healthy control (Control) group. Flow cytometry was utilized to determine cell apoptosis. Proteins and messenger RNAs (mRNAs) were estimated by Western blot and quantitative reverse transcription PCR (qRT-PCR), respectively.
Results: LINC00673 was downregulated in plasma samples of DR patients (n=60) in comparison with the healthy controls (n=60) and negatively correlated with p53 only across DR patients but not across the healthy controls. In retinal pigment epithelial cells (RPECs), high glucose treatment downregulated LINC00673. Moreover, LINC00673 overexpression downregulated p53 and decreased RPEC apoptosis, while LINC00673 silencing upregulated p53 and increased RPEC apoptosis. In addition, p53 overexpression reduced the effects of LINC00673 overexpression.
Conclusion: LINC00673 is downregulated in DR patients and regulates RPEC apoptosis via negatively regulating p53.
Keywords: diabetic retinopathy, LINC00673, p53, apoptosis
Introduction
Diabetes mellitus is a common metabolic disorder worldwide. In 2017, more than 425 million people suffered from diabetes mellitus, and this number will rise to more than 629 million by 2045 due to people’s lifestyle changing and the aging population growing.1,2 Diabetes mellitus causes a series of complications that affects almost all major organs in the human body, such as the lungs, eyes, feet, and hands.3 As one of the most common types of diabetic complications, diabetic retinopathy (DR) is the leading cause of blindness among adults of working age.4 Although the progression of DR can be suppressed by proper treatment, such as blood glucose control, blindness will inevitably occur in a considerable portion of DR patients even after active treatment.5 Therefore, a better understanding of the pathogenesis of DR is needed to develop novel therapeutic approaches.
As a cell apoptosis mediator, p53 signaling plays a pivotal role in the development of diabetic complications, including DR.6 In DR, high-glucose treatment activates p53 to induce cell apoptosis, thereby aggregating disease conditions.6 In fact, inhibition of p53 is considered a promising approach to treat diabetic complications by inhibiting cell apoptosis.7,8 In some cases, p53 signaling is transduced via interacting with long noncoding RNAs (>200nt, lncRNAs),9,10 which have important roles in diabetic complications.11 In DR, lncRNAs interact with multiple signaling pathways to participate in nearly all aspects of the development and progression of DR.11 Regulating the expression of certain key lncRNAs may contribute to the recovery of DR.11 Meng et al showed that lncRNA LINC00673 suppresses the progression of papillary thyroid carcinoma by downregulating p53.12 Therefore, it is reasonable to hypothesize that LINC00673 may also participate in DR by interacting with p53. This study was therefore performed to test this hypothesis.
Materials and Methods
Study Subjects and Plasma
The present study included 3 groups of participants, namely DR group, diabetes (DB) group, and healthy control (Control) group. DR group included 60 patients with DR (34 males and 26 females, 38 to 66 years, 50.1±6.7 years), DB group included 60 diabetic patients without obvious complications (34 males and 26 females, 37 to 66 years, 49.7±6.8 years), and Control group included 60 healthy volunteers (34 males and 26 females, 36 to 67 years, 49.9±6.9 years) from the Physiological Health Center. All participants were admitted to the Lianyungang Second People’s Hospital between December 2016 and December 2018. The inclusion criteria were 1) all patients were newly diagnosed and 2) no therapies were performed. The exclusion criteria were 1) other clinical disorders were observed and 2) patients received any therapies within 3 months before admission. All patients were educated with experimental principle and signed informed consent. This study passed the review of the Ethics Committee of the aforementioned hospital. Table 1 lists the basic clinical information of the 3 groups of patients.
 |
Table 1 Basic Clinical Characters of Participants in the 3 Groups at the First Diagnose |
Serum Collection
Blood (5 mL) was extracted from each participant before the initiation of any therapies, collected in ethylenediaminetetraacetic acid disodium salt (EDTA) tubes, and centrifuged at 1200 g for 16 min to collect the supernatant (plasma).
Cells and Transfections
Human retinal RPEC cell line h1RPE7 and ARPE-19 were purchased from Sigma-Aldrich (USA) and cultured in DMEM: F12 media containing 10% FBS at 37 °C in a humidified incubator with 5% CO2.
LINC00673 and p53 expression pcDNA3 vectors were from GenePharma (Shanghai, China). Negative control (NC) siRNA and LINC00673 siRNA were from Sangon (Shanghai, China). h1RPE7 cells at 70–80% confluency (about 105 cells) were transfected with 10 nM LINC00673 and p53 expression pcDNA3 vector (empty pcDNA3 vector as NC group) or 45 nM LINC00673 siRNA (NC siRNA as NC group) using Lipofectamine 2000 reagent (Invitrogen, USA). Control cells were untransfected cells. Subsequent experiments were performed at 24h of post-transfection.
Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
Total RNAs were extracted from 0.3 mL plasma or 105 h1RPE7/ARPE-19 cells using Trizol reagent kit (Invitrogen, USA). For high glucose treatment, h1RPE7/ARPE-19 cells were treated with 5, 10, 30, and 60 mM D-glucose for 24h before total RNA extraction. A total of 0.5 µg RNA from each sample were treated with DNase I and subjected to reverse transcriptions using Quantitect Reverse Transcription Kit (Qiagen, Shanghai, China). A total of 1 µL cDNA template were added into a PCR reaction system with 10 nM primers, 6 ul SYBR® Green Master Mix (Bio-Rad, USA), and 5 µl water. The pPCRs were performed for 35 cycles of annealing at 55 °C and the extension at 72°C in a CFX96 Touch Deep Well RT-PCR System (Bio-Rad, Hercules, CA). LINC00673 and p53 mRNA expression was normalized to 18S rRNA, and the fold changes in mRNA expression levels were calculated using the 2−ΔΔCt method. Aliquots of the PCR products were separated on 1.5% agarose gels, and PCR fragments were visualized by ethidium bromide staining. All the primer sequences are available in Table 2.
 |
Table 2 Primer Sequences Used in PCR |
Enzyme-Linked Immunosorbent Assay (ELISA)
Serum p53 levels were detected using human p53 pre-coated ELISA kits (ab46067, Abcam) in accordance with the manufacturer’s instruction. The absorbance at 450 nm of each well was collected using a microplate reader (Thermo, Massachusetts). The standard diluents were used for generating the standard curve. All samples were measured in triplicate. The inter and the intra-assay coefficients derived from 6 to 8 samples were 22.5±1.67 ng/µL (mean±SD, CV=6.2%) and 51.6±0.45 ng/µL (mean±SD, CV=3.7%), respectively. Plasma p53 levels were normalized to pg/mL with the minimum detectable level of <1.5 pg/mL.
Cell Apoptosis Analysis
h1RPE7 and ARPE-19 cells were harvested at 24h after transfections. Under the conditions of 37 °C and 5% CO2, h1RPE7 cells were cultured in media containing 30 and 60 mM D-glucose for 24h with 3 biological replicates per treatment. Each replicate included 2 mL of cell suspensions containing 6×104 cells. After D-glucose treatment, h1RPE7 and ARPE-19 cells were digested with 0.25% trypsin and suspended to a density at 5×106 cells/mL, followed by staining with Annexin V-FITC (Dojindo, Japan) for 15 min and propidium iodide (Dojindo, Japan) for 5 min and in the dark. After removal of the staining solution, apoptotic cells were detected and analyzed using flow cytometry (FACSCanto II, BD Bioscience, USA) and Flowjo software.
Western Blot
h1RPE7/ARPE-19 cells were counted, and cell pellets containing 2×105 cells were resuspended in 1 mL RIPA solution (Cat# C500007-0010, Sangon) to perform total protein extractions. Protein samples were boiled in water for 5 min, followed by separation with 10% SDS-PAGE gel electrophoresis. The separated proteins were transferred onto poly(vinylidene fluoride) (PVDF) membranes, followed by incubation with rabbit polyclonal primary antibodies against GAPDH (ab9485, 1:800, Abcam) and p53 (ab131442, 1:800; Abcam) for at least 12 h at 4°C. The membranes were further incubated with horseradish peroxidase-labeled IgG secondary antibody (1:800, MBS435036, MyBioSource) for 2h at 22 °C. Electrochemiluminescence (ECL) substrate (Sigma-Aldrich, USA) was dropped onto the PVDF membranes to produce signals. Data were analyzed using Image J v1.46 software.
Statistical Analysis
Each experiment was repeated 3 times to calculate the mean values. Differences among different participant groups and among different cell groups were explored using analysis of variance (ANOVA, one-way) and Tukey’s test. Correlations were analyzed using linear regression. Comparisons of clinical data among the 3 groups were performed by Chi-squared t-test. A p < 0.05 was statistically significant.
Results
LINC00673 and p53 Showed Opposite Expression Pattern in DR
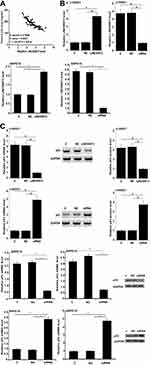
Serum LINC00673 and p53 levels of the 3 groups of participants were detected by RT-qPCR and ELISA, respectively. Plasma LINC00673 levels (Figure 1A) were significantly lower, and p53 levels were significantly higher (Figure 1B) in DR group than in DB and Control groups (p < 0.05). In addition, plasma LINC00673 levels (Figure 1A) were significantly lower, and p53 levels were significantly higher (Figure 1B) in DB group than in Control group (p < 0.05). Moreover, analysis of the correlations between LINC00673 and p53 showed that plasma LINC00673 and p53 levels were inversely and significantly correlated in DR patients (Figure 2A).
High Glucose Environment Downregulated LINC00673 in h1RPE7 and ARPE-19 Cells
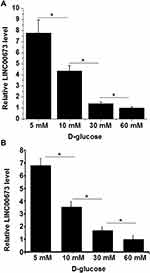
h1RPE7 and ARPE-19 cells were treated with D-glucose at doses of 5 (physiological level), 10, 30 and 60 mM for 24h, and LINC00673 expression was detected by RT-qPCR. It was observed that D-glucose downregulated LINC00673 in a concentration-dependent manner in vitro (Figure 3, p < 0.05).
LINC00673 Negatively Regulated p53 in h1RPE7 and ARPE-19 Cells
LINC00673 expression vector and siRNA were transfected into h1RPE7 cells and ARPE-19 cells. At 24h following transfections, LINC00673 expression was significantly altered at 24h after transfections comparing to C and NC groups (Figure 2B, p < 0.05). In addition, compared to C and NC groups, LINC00673 overexpression downregulate, while LINC00673 siRNA silencing upregulated p53 (Figure 2C, p < 0.05). LINC00673 may negatively regulate p53 expression as a competing endogenous RNA (ceRNA). We searched the LncBase database and miRWalk database for ceRNA analysis on LINC00673 and p53. We selected the top 10 target miRNAs of LINC00673 with the most associations and used Cytoscape to draw the ceRNA network map of LINC00673, as shown in Supplementary Materials.
LINC00673 Inhibited the Apoptosis of h1RPE7 and ARPE-19 Cells Under High Glucose Environment via p53
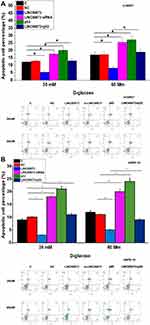
h1RPE7 and ARPE-19 cells were treated with 30 and 60 mM D-glucose for 24h, followed by the analysis of cell apoptosis. Compared to C and NC groups, LINC00673 overexpression decreased while LINC00673 siRNA silencing increased the apoptotic rate of RPECs (Figure 4, p < 0.05). In addition, p53 overexpression reduced the effect of LINC00673 overexpression (Figure 4, p < 0.05).
Discussion
In the present study, we investigated the role of LINC00673 in DR and found that LINC00673 was downregulated in DR, and LINC00673 overexpression might prevent the apoptosis of retinal pigment epithelial cells by downregulating p53.
The expression pattern and functionality of LINC00673 have been widely studied in cancer biology.12,13 However, the involvement of this lncRNA in other human diseases is still unknown. In this study, we observed downregulated LINC00673 in DR and DB patients compared to healthy controls. In addition, LINC00673 expression levels were also significantly lower in DR patients than in DB patients. Moreover, high glucose treatment also downregulated LINC00673 in retinal pigment epithelial cells. Therefore, it is reasonable to hypothesize that LINC00673 is downregulated with the development of diabetes, and the further downregulation of LINC00673 is accompanied by the occurrence of DR, or LINC00673 downregulation contribute to the development of DR.
Retinal pigment epithelium is the blood-retinal barrier.14 During the development of DR, retinal pigment epithelial cell apoptosis is induced, and barrier functions of the retinal pigment epithelium are seriously affected.14,15 Therefore, inhibition of retinal pigment epithelial cell apoptosis is critical to maintaining normal eye functions.14,15 The apoptosis of retinal pigment epithelial cells induced by high glucose metabolism can be regulated by lncRNAs. LncRNA BDNF-AS downregulates BDNF to regulate the apoptosis of retinal pigment epithelial cells induced by high glucose.16 In another study, Zhou et al reported that downregulation of lncRNA NKILA played a protective role in retinal pigment epithelial cells under hypoxic conditions.17 Inactivation of p53 signaling prevents the apoptosis of retinal pigment epithelial cells induced by oxidative damage.18 It has been reported that LINC00673 interacts with EZH2 and DNMT1 to downregulate p53 in papillary thyroid carcinoma.12 In this study, we proved that LINC00673 also negatively regulated p53 in retinal pigment epithelial cells, and p53 downregulation by LINC00673 suppressed the apoptosis of retinal pigment epithelial cells under high glucose treatment. Therefore, LINC00673 overexpression might improve the conditions of DR.
Conclusion
LINC00673 was downregulated in DR, and LINC00673 overexpression may inhibit the apoptosis of retinal pigment epithelial cells in a high glucose environment by downregulating p53.
Availability of Supporting Data
The data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants but available on request from the corresponding author.
Ethical Approval and Consent to Participate
All patients were educated with the experimental principle and signed informed consent. All procedures were approved by the Ethics Committee of Lianyungang Second People’s Hospital and operated in keeping with the standards set out in the Announcement of Helsinki and Laboratory Guidelines of Research in China.
Funding
There is no funding to report.
Disclosure
The authors declare that they do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
References
1. Karuranga S, Fernandes JDR, Huang Y, Malanda B; International Diabetes Federation, editors. IDF Diabetes Atlas.
2. Toi PL, Anothaisintawee T, Chaikledkaew U, Briones JR, Reutrakul S, Thakkinstian A. Preventive role of diet interventions and dietary factors in type 2 diabetes mellitus: an umbrella review. Nutrients. 2020;12(9):2722. doi:10.3390/nu12092722
3. Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54(6):1615–1625. doi:10.2337/diabetes.54.6.1615
4. Fong DS, Aiello L, Gardner TW, et al. Retinopathy in diabetes. Diabetes Care. 2004;27(Suppl 1):S84–S87. doi:10.2337/diacare.27.2007.S84
5. Tracey ML, McHugh SM, Fitzgerald AP, Buckley CM, Canavan RJ, Kearney PM. Trends in blindness due to diabetic retinopathy among adults aged 18–69 years over a decade in Ireland. Diabetes Res Clin Pract. 2016;121:1–8. doi:10.1016/j.diabres.2016.08.016
6. Kung CP, Murphy ME. The role of the p53 tumor suppressor in metabolism and diabetes. J Endocrinol. 2016;231(2):R61–R75. doi:10.1530/JOE-16-0324
7. Kundu N, Domingues CC, Chou C, et al. Use of p53-silenced endothelial progenitor cells to treat ischemia in diabetic peripheral vascular disease. J Am Heart Assoc. 2017;6(4). doi:10.1161/JAHA.116.005146
8. Gu J, Wang S, Guo H, et al. Inhibition of p53 prevents diabetic cardiomyopathy by preventing early-stage apoptosis and cell senescence, reduced glycolysis, and impaired angiogenesis. Cell Death Dis. 2018;9(2):82. doi:10.1038/s41419-017-0093-5
9. Grossi E, Sánchez Y, Huarte M. Expanding the p53 regulatory network: lncRNAs take up the challenge. Biochim Biophys Acta. 2016;1859(1):200–208. doi:10.1016/j.bbagrm.2015.07.011
10. Wu Z, Wu P, Zuo X, et al. LncRNA-N1LR enhances neuroprotection against ischemic stroke probably by inhibiting p53 phosphorylation. Mol Neurobiol. 2017;54(10):7670–7685. doi:10.1007/s12035-016-0246-z
11. Leung A, Natarajan R. Long noncoding RNAs in diabetes and diabetic complications. Antioxid Redox Signal. 2018;29(11):1064–1073. doi:10.1089/ars.2017.7315
12. Meng XF, Zhao LY, Chu XF. LncRNA LINC00673 inhibits p53 expression by interacting with EZH2 and DNMT1 in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(5):2075–2083.
13. Huang M, Hou J, Wang Y, et al. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. 2017;25(4):1014–1026. doi:10.1016/j.ymthe.2017.01.017
14. Simó R, Villarroel M, Corraliza L, Hernández C, Garcia-Ramírez M. The retinal pigment epithelium: something more than a constituent of the blood-retinal barrier–implications for the pathogenesis of diabetic retinopathy. J Biomed Biotechnol. 2010;2010:190724. doi:10.1155/2010/190724
15. Xia T, Rizzolo LJ. Effects of diabetic retinopathy on the barrier functions of the retinal pigment epithelium. Vision Res. 2017;139:72–81. doi:10.1016/j.visres.2017.02.006
16. Li Y, Xu F, Xiao H, Han F. Long noncoding RNA BDNF-AS inversely regulated BDNF and modulated high-glucose induced apoptosis in human retinal pigment epithelial cells. J Cell Biochem. 2018;119(1):817–823.
17. Zhou Q, Zhou L, Qian J, Yuan ZL, Chen ZJ. NKILA inhibition protects retinal pigment epithelium cells from hypoxia by facilitating NFκB activation. Biochem Biophys Res Commun. 2018;503(4):3134–3141. doi:10.1016/j.bbrc.2018.08.105
18. Gong L, Liu F, Xiong Z, et al. Heterochromatin protects retinal pigment epithelium cells from oxidative damage by silencing p53 target genes. Proc Natl Acad Sci USA. 2018;115(17):E3987–E3995. doi:10.1073/pnas.1715237115
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.