Back to Journals » OncoTargets and Therapy » Volume 12
LncRNA LINC00461 Promotes Colorectal Cancer Progression via miRNA-323b-3p/NFIB Axis
Authors Yu H, Ma J, Chen J, Yang Y, Liang J, Liang Y
Received 27 August 2019
Accepted for publication 4 November 2019
Published 17 December 2019 Volume 2019:12 Pages 11119—11129
DOI https://doi.org/10.2147/OTT.S228798
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Hairong Yu,1,* Jianguo Ma,2,* Jianshuang Chen,1 Yang Yang,1 Jianjing Liang,3 Yulong Liang4
1Functional Experiment Center, Chengde Medical College, Chengde 067000, People’s Republic of China; 2Department of Urology, The Third Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China; 3Medical Department of Hebei University, Hebei University, Baoding, Hebei, People’s Republic of China; 4Department of General Surgery, The Third Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yulong Liang Tel +8618533112938
Email [email protected]
Background: LncRNA LINC00461 has been reported to play crucial regulatory roles in a variety of biological processes, including cell migration, cell invasion and cancer progression. However, its biological role in colorectal cancer (CRC) is completely unknown. The aim of our study was to explore the function of LINC00461 on CRC cells and the underlying mechanism.
Materials and methods: CRC tumor tissues and cell lines derived from hospital and corporation. The expression level of LINC00461 in CRC tissues and cell lines were analyzed by quantitative real-time PCR (qRT-PCR). The effect of LINC00461 on cell proliferation, colony formation, migration and invasion were detected by CCK-8 assay, colony formation and transwell assay, respectively. In addition, cell apoptosis was analyzed by flow cytometry, and the role of LINC00461 on tumor growth was investigated by tumor xenografts in nude mice. The targets of LINC00461 were predicted by starBase v3.0 and confirmed by a dual-luciferase reporter system. The expression level of transcription factors of nuclear factor I B (NFIB), p21 and CDK2 was determined by Western blot or qRT-PCR. The NFIB expression levels in CRC tissues and mice tumors were analyzed by immunofluorescence assay (IHC).
Results: We found that the expression of LINC00461 was significantly overexpressed in CRC tissues and different cell lines, and the high level of LINC00461 expression was associated with poor overall survival. Downregulation of LINC00461 expression significantly suppressed the proliferation, migration and invasion of CRC cells and promoted cell apoptosis. We also found that LINC00461 could directly interact with miR-323b-3p. In addition, LINC00461 significantly increased the expression NFIB and CDK2, but, p21 was inhibited. Finally, we found that the growth of tumors in nude mice was suppressed upon LINC00461 deletion.
Conclusion: We demonstrated that LINC00461 may play an oncogenic role in CRC cells through NFIB signaling pathway by targeting miR-323b-3p. Our report showed that LINC00461 may be a prognostic biomarker and candidate therapeutic target for CRC.
Keywords: LINC00461, colorectal cancer, miR-323b-3p, NFIB
Introduction
As a kind of human cancer, colorectal cancer is the second cause about cancer-related death in western countries.1 Simultaneously, in China, death from colorectal cancer is also the fifth cause about cancer-related death2 due to the lack of tumor diagnosis method to rapid cancer progression.3 In addition, high mortality of CRC is a lack of availability of adequate prognostic biomarkers, high degree of metastasis capacity, poor prognosis and recurrence. Report showed that about 90% of early patients of CRC have a chance to survive by surgery. But, many patients have already diagnosed at advanced stages.4 Despite chemotherapy, surgery and even immunotherapy are used to treat patients with CRC in current clinical treatments, the poor prognosis is not eliminated in patients with advanced disease.5–7 That is why it is very necessary that further clarify the potential pathogenesis and explore predictive markers to improve the survival rate and prognosis of CRC patients.
Long non-coding RNAs (lncRNAs) are made up of noncoding RNA with a length of more than 200 nucleotides, but no protein encoding function.8 Accumulating evidence revealed that the dysregulation of lncRNAs plays significant roles in many physiological and pathological processes of human diseases, especially in cancers. Some reports showed that lncRNAs are related to pathogenesis of cancers by promoting or inhibiting the onset of cancers, the dysregulation of lncRNAs expression can affect the cycle, proliferation, growth, apoptosis, metastasis and invasion of cancer cells by mediating epigenetic modifications and regulating transcriptional activities.9 In recent years, studies about lncRNAs have attracted an ever-increasing number of attention due to its function on microRNA (miRNA). LncRNA can regulate the expression of protein by targeting miRNA.9,10 LINC00461 was a newly identified LncRNA, studies indicated that the level of LINC00461 expression was upregulated in glioma, hepatocellular carcinoma and breast cancer.11–13 But, the biological function of LINC00461 in CRC remains unclear.
In our work, results demonstrated that the expression of LINC00461 was substantially upregulated in tissue and cells of CRC, and it is related with the poor overall survival. We further explored the roles of LINC00461 on the CRC cells in vitro, results dedicated that LINC00461 can promote the cell proliferation, invasion and migration, but inhibit cell apoptosis by targeting miR-323b-3p that can target NFIB and regulating the expression of p21 and CDK2. The growth of CRC cells was significantly inhibited due to the silence of LINC00461 in vivo. Taken together, our study reveals that LINC00461 can be served as a candidate biomarker for CRC diagnosis and treatment.
Materials and Methods
Patients and Samples
Colorectal cancer tumor tissues of human and corresponding adjacent normal tissues were obtained from Peking Union Medical College. All patients did not undergo any treatment and provided written informed consent. This study was approved by the Ethics Committee of Chengde Medical Collage and Peking Union Medical College. All experiments involving human CRC tissues were performed in accordance with the Declaration of Helsinki.
Cell Culture
The human normal colonic epithelial cell NCM460, colonic cancer cell lines (DLD-1, RKO, HT29, and SW480) and HEK293T were purchased from the ScienCell Research Laboratories. All cells were cultured in DMEM medium (Gibco, USA) containing 10% FBS (Gibco, USA), and placed in a cell culture incubator with 5% CO2 at 37°C.
RNA and qRT-PCR
In order to obtain RNA, cells and tumor issues were lysed by TRIzol (Invitrogen, USA) and reverse transcription was performed by a reverse transcription kit (Invitrogen, USA). SYBR-Green PCR Master Mix Kit (Takara, Japan) is used to perform Quantitative PCR on an ABI 7500 system (Applied Biosystems, Foster City, USA). The sequences of real-time PCR primers were presented as follows:
LINC00461: Forward (5ʹ-3ʹ) GACATTTACGCCACAACCCACG;
Reverse (5ʹ-3ʹ): AGACAGACCCTCAGATTCCCCA.
U6: Forward (5ʹ-3ʹ) CTCGCTTCGGCAGCACA;
Reverse (5ʹ-3ʹ) AACGCTTCACGAATTTGCGT.
NFIB: Forward (5ʹ-3ʹ) TGAGGCAGCTTCACCTACAG;
Reverse (5ʹ-3ʹ) AGGATGGGTCTCTTGGGCTTA.
GAPDH: Forward (5ʹ-3ʹ) GTCAACGGATTTGGTCTGTATT;
Reverse (5ʹ-3ʹ) AGTCTTCTGGGTGGCAGTAT.
hsa-miR-323b-3p: Forward (5ʹ-3ʹ) TGCGGCCCAATACACGGTCGACC;
Reverse (5ʹ-3ʹ) CCAGTGCAGGGTCCGAGGT.
Cell Transfection
si-LINC00461, miR-323b-3p mimic, miR-323b-3p inhibitor, and their negative control RNA (si-NC) were constructed and purchased from Jikai Chemical Technology (Shanghai, China). In order to overexpress LINC00461, full-length LINC00461 was cloned into the pcDNA3.1 vector (Genechem, Shanghai, China). Cells were transfected with si-NC, si-LINC00461, pcDNA3.1, pcLINC00461, miR-323b-3p mimics, miR-NC, miR-323b-3p inhibitor and inhibitor-NC using the Lipofectamine 2000 reagent (Invitrogen, USA), respectively. The transfection efficiency was analyzed by qRT-PCR assay after 48 h post-transfection.
Cell Viability Analysis
CRC cells were plated in 96-well plates (2000 cells/well). Then, 10 μL of the Cell Counting Kit-8 (Dojindo, Japan) was added into each well for 1–4 h. Optical density (OD) values at 450 nm were measured by a microplate reader (Thermo Scientific, USA). The cell viability was calculated according to the manufacturer’s instruction.
Colony Formation Assay
DLD-1 cells first were transfected for 24 h, and then cultured in six-well plates (1×103 cells/well) for 2 weeks. Colonies were fixed with 4% of paraformaldehyde for 10 min, stained with 1% crystal violet, and washed twice with PBS. Finally, cell colonies were imaged and counted.
Cell Migration and Invasion
To assess the cell migration and invasion capability, transwell assay was performed, DLD-1 cells were transfected with different plasmid or si-RNA. Cells were suspended by medium without serum, 5×104 cells were seeded in the upper chamber. Cultured medium with 20% serum was placed in the lower chamber. Then, noninvading and nonmigrating cells were cleared after 48 h. Remaining cells were fixed with 4% of paraformaldehyde for 15 min, the membranes were stained with 0.1% crystal violet solution for 30 min. Then, the stained cells were captured using a microscope and counted.
Dual-Luciferase Reporter Assay
We seeded DLD-1 cells in a 96-well plate (5000 cells/well). Then, cells were co-transfected with luciferase reporter plasmids of LINC00461-wild type or LINC00461-mutant type and miR-323b-3p mimics or miR-NC using RNAiMAX Reagent (Thermo Scientific, USA) according to the reagent instruction. Luciferase activities were detected by a microplate reader of a multi-wavelength measurement system (Bio-Rad, USA) at 48 h post-transfection. Similarly, NFIB-wild type or NFIB-mutant type luciferase reporter plasmids were co-transfected with miR-323b-3p mimics or miR-NC into HEK293T cells, luciferase activities were measured as previously described.
Flow Cytometric Analysis of Apoptosis
To evaluate the effect of LINC00461 to cell apoptosis, the cell apoptosis assay was performed by a FITC Annexin V Apoptosis Detection Kit (BD Biosciences, USA) according to the manufacturer’s instruction. DLD-1 cells transfected with si-NC or si-LINC00461 for 48 h, then cells were collected and stained with Annexin V for 10 min and Propidium Iodide (PI) for 5 min in the dark. Last, the percentage of apoptotic cells was measured using a BD FACSCalibur flow cytometer (BD Biosciences, USA).
Western Blot
Cells were lysed with RIPA lysis buffer (Solarbio), protein samples were subjected to Western blot with primary antibody for anti-NFIB (Abcam, UK), anti-p21 (Abcam, UK), anti-CDK2 (Abcam, UK), anti-GAPDH (Abcam, UK) and secondary antibody.
Tumor Xenografts in Nude Mouse
All animal experiments in this study were performed according to guidelines of the NIH for the care and use of laboratory animals and approved by the Animal Ethics Committee of Chengde Medical Collage. si-NC or si-LINC00461 was transfected into DLD-1 cells, and cells were subcutaneously injected into female BALB/c nude mice (4-week old, n=10). Tumor size was measured after injection and growth curves were plotted. Tumor volume was calculated according to the formula: Volume = Length×Width×Width×1/2. At post-injection 28 days, mice were sacrificed, and other experiments were performed.
Immunohistochemistry
In order to detect the expression of NFIB in tumor tissues, we performed immunohistochemistry assays (IHC) according to previously described method.14 The primary and secondary antibodies in Western blot were also used for Immunohistochemistry.
Lung Metastatic Mouse Model
DLD-1 cells were transfected with si-NC or si-LINC00461 and injected into nude mice via tail vein (4×106 cells/200 µL PBS/mouse). All mice were sacrificed after 40 days, lung tissues were harvested and imaged, and the number of metastatic lesions in the lungs was counted to evaluate the role of LINC00461 on CRC lung metastasis.
Statistical Analysis
All data were presented as mean ± SD. Differences between two experimental groups were analyzed by a paired t-test. P < 0.05 is considered to have significant statistical differences.
Results
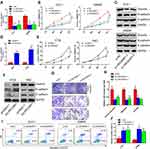
LINC00461 Was Upregulated in Human CRC Tissues and Cell Lines
To investigate the role of LINC00461 in CRC, the expression level of LINC00461 was examined from 30 patients with CRC. Quantitative PCR showed that higher expression of LINC00461 in the CRC tumour tissues instead of adjacent tissues (Figure 1A). The expression of LINC00461 was also analyzed in CRC cell lines (RKO, HT29, DLD-1 and SW480) and a normal cell line (NCM460). We found that the LINC00461 expression level was highly increased in cancer cells (Figure 1B). Furthermore, we found that LINC00461 expression was positively along with the progress of cancer, the expression level of LINC00461 in III/IV stages was obviously higher than I/II stages (Figure 1C). Meanwhile, we further analyzed the survival rate of patients. Result showed that downregulation of LINC00461 had higher survival rates than high LINC00461 expression (Figure 1D). Those results showed that LINC00461 might be related to CRC progress.
LINC00461 Affects Proliferation, Colony Formation, Migration and Invasion of CRC Cell in vitro
Next, we have further studied the function of LINC00461 on CRC cells, si-NC, si-LINC00461 or si-LINC00461-1 was transfected into DLD-1 and SW480 cells. Correspondingly, pcDNA3.1 or pcLINC00461 overexpression plasmids were transfected into RKO and HT29 cells. The transfection efficiency was confirmed by qRT-PCR at 48 h post-transfection (Figure 2A and D). Our results showed that the proliferation capacity of cells was significantly suppressed after transfected with si-LINC00461 as compared with control group (Figure 2B). As we expected, the overexpression of LINC00461 prominently promoted the proliferation capacity of cells (Figure 2E). Simultaneously, we further explored the role of LINC00461 to cells on mesenchymal features. Results indicated that the expression of Vimentin and N-cadherin was reduced, but the expression of E-cadherin was increased when cells transfected with si-LINC00461 (Figure 2C). On the contrary, these appearances in Figure 2C were reversed in cells transfected with LINC00461 overexpression plasmids (Figure 2F). Next, colony formation and transwell assays showed that the downregulation of LINC00461 expression level extremely suppressed colony formation, migration and invasion capacities of CRC cells (Figure 2G and H). Finally, the function of LINC00461 to cell apoptosis was explored with flow cytometry. We found that cell apoptosis rates were substantially increased when cells were transfected with si-LINC00461 (Figure 2I and J). In summary, these results indicated that LINC00461 can regulate cell colony formation, apoptosis, migration and invasion.
LINC00461 Directly Interacts with miR-323b-3p in CRC
Many studies showed that lncRNAs can be combined with miRNAs with complementary base pairing way. To predict potential targets of LINC00461, the potential miRNAs were predicted by starBase v3.0. A potential complementary binding site of miR-323b-3p and LINC00461 as shown in Figure 3A. To prove it, luciferase reporter experiments were carried out. DLD-1 cells were transfected with luciferase reporter plasmids and miR-323b-3p mimics. Results showed that luciferase activity decreased for cells transfected with wild-type vector. But the luciferase activity of mutant-type vector no affected (Figure 3B). In addition, we also detected the expression of miR-323b-3p in tumor tissues of CRC and cell lines. Our results showed that the expression of miR-323b-3p was decreased in tumor tissues of CRC and cell lines (Figure 3C and D). Simultaneously, spearman correlation test demonstrated that LINC00461 was negatively correlated with miR-323b-3p in tissues of CRC (Figure 3E). Furthermore, we found that the downregulation of LINC00461 leads to the increasement of miR-323b-3p in DLD-1 and SW480 cells (Figure 3F). While the expression of LINC00461 was down-regulated when miR-323b-3p mimics were transfected into HT29 and RKO cells (Figure 3G). In addition, we demonstrated that cell proliferation capacity was significantly suppressed in DLD-1 cells transfected with miR-323b-3p mimics, the opposed results were found in DLD-1 cells transfected with miR-323b-3p inhibitor (Figure 3H). Rescue assay also indicated LINC00461 and miR-323b-3p can reciprocally reverse the respective inhibition effect for DLD-1 cell proliferation (Figure 3I and J). These data suggest that LINC00461 can directly interact with miR-323b-3p and correlated negatively with miR-323b-3p.
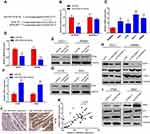
miR-323b-3p Inhibits CRC Progression by Targeting NFIB
To identify potential target genes of miR-323b-3p, bioinformatic analysis starBase v3.0 predicted that miR-323b-3p directly targets NFIB (Figure 4A). HEK293T cells were transfected with NFIB luciferase reporter plasmids and miR-323b-3p mimics. Results indicated that overexpression of miR-323b-3p highly reduced fluorescence intensity in cells transfected with wild-type vector (Figure 4B). In addition, qRT-PCR indicated that the expression of NFIB was increased in CRC cell lines (Figure 4C). Furthermore, Western blot analysis and qRT-PCR also has proven that the expression of NFIB was reduced in cells transfected with miR-323b-3p mimics (Figure 4D and E). But inhibitory effect was abolished in cells transfected with miR-323b-3p inhibitor (Figure 4F and G). Next, in order to further explore the roles of LINC00461 on NFIB expression, the CRC cells were transfected with si-LINC00461 or pcLINC00461. Results showed that the down expression of LINC00461 has significantly decreased NFIB expression, we also observed the expression of p21 (a direct target of NFIB15) was induced, CDK2 (cyclin-dependent kinase 2, combination of p21 and CDK2 leads to cell cycle arrest.16) was suppressed upon LINC00461 deletion (Figure 4H). On the contrary, NFIB expression was increased in cells with high LINC0046 expression, however, p21 and CDK2 presented the opposite results as compared with Figure 4H upon LINC00461 overexpression (Figure 4I). Tissue immunofluorescence assay also showed that NFIB was induced in tumor tissues of CRC patients (Figure 4J). Spearman correlation test has also indicated the positive correlation between LINC00461 and NFIB (Figure 4K). Collectively, these data indicated that NFIB can server as a target of miR-323b-3p.
Downregulation of LINC00461 Inhibits Tumor Growth
Lastly, the function of LINC0046 on tumor was investigated in vivo. Nude mice were subcutaneously injected with DLD-1 cells transfected with si-NC or si-LINC00461. We found that the growth of tumor in si-LINC00461 group was slower than si-NC group (Figure 5A). The weight of the tumor was measured at 28 days post-injection. Results indicated that tumor weight in si-LINC00461 group was obviously less than si-NC group (Figure 5B). The high expression of NFIB was also found by IHC in tumor issues of mice transfected with si-NC instead of si-LINC00461 (Figure 5C). Interestingly, tumor metastases assay also indicated the knockdown LINC00461 inhibited the lung metastasis of CRC (Figure 5D and E). These data demonstrated that downregulation of LINC00461 obviously suppressed CRC progression in vivo. At last, the schematic illustration of LINC00461 regulating miR-323b-3p/NFIB axis showed in Figure 5F.
Discussion
A lot of researches have indicated that lncRNAs are closely involved in the human diseases. As oncogenes or cancer suppressor genes, the dysregulation of lncRNAs expression was closely related with the progression of cancers.17,18 LncRNAs with significant roles in growth characteristics of cancer cells can regulate the epithelial-mesenchymal transition (EMT), metastasis, apoptosis, migration and invasion.19–21 In this study, we found that LINC00461 expression was significantly increased in tumor tissues and cell lines of CRC. In addition, the poor OS of patients with CRC was correlated with high LINC00461 expression. The proliferation, migration and invasion capacities were inhibited in CRC cells transfected with si-LINC00461. However, the effect was significantly reversed by the upregulation of LINC00461.
Some reports have demonstrated that LINC00461 was associated with the progress of various cancers by targeting miRNA. For example, Ji et al reported that LINC00461 have a high expression level in hepatocellular carcinoma, it positively correlated with advanced stage, poor prognosis and metastasis.12 Dong et al reported that LINC00461 exerted a promoted effect in breast cancer by enhancing cell migration and invasion.13 Interestingly, Gao et al showed that LINC00461 was related to macular thickness at genome-wide, characterized by elevated expression of LINC00461 in the retina, which indicated that LINC00461 also exerted function in other human disease.22 Deng et al demonstrated that high LINC00461 expression dramatically promoted cell proliferation and suppressed cell apoptosis in multiple myeloma in the way of regulating miR-15a/miR-16 on BCL-2.23 Yang et al indicated that LINC00461 enhanced the progress of glioma by mediating AMAPK/ERK and PI3K/AKT signaling pathways.11 In this work, we found that cell viability, invasion and migration capacities were obviously suppressed for the LINC00461 downregulation in vitro and in vivo, and cell apoptosis was promoted.
To explore the potential roles of LINC00461 on CRC, starBase v3.0 is used to predict the targets of LINC00461. We found that miR-323b-3p could be a target and based on its biological function in cancers. Xie et al reported that miR-323b-3p can regulate osteosarcoma (OS). TGFBR3, a target of miR-323b-3p, was inhibited at the early stage of OS. But, TGFBR3 was promoted by miR-323b-3p in the advanced stage of OS. So, miR-323b-3p plays a dual role in different stages of OS by mediating TGF-β signaling pathway.24 In addition, the expression of miR-323b-3p is significantly increased and closely correlated with metastasis and poor prognosis of lung adenocarcinoma, and genomic hypomethylation.25 miR-323b-3p was also reported to may be associated with lupus nephritis.26 However, study showed that down-regulation of miR-323b-3p expression may be related to the progress of diffuse large B-cell lymphoma.27 NFIB was reported to regulate many cellular gene expression.28 Reports showed that NFIB plays a vital role in tumor progression. Liu et al showed that NFIB exerted positive role in the progress of triple-negative breast cancer by promoting cell survival.15 Campbell et al demonstrated that NFIB and YBX1 have affected the breast cancer progress by binding ERα, leading to FGFR2 to modulate estrogen responsiveness.29 In addition to the descriptions above, NFIB also be reported to regulate other cancer progressions in glioma, glioblastoma and osteosarcoma.14,30–32 But the relationship between NFIB and miR-323b-3p is still unknown in CRC. In our study, we indicated that LINC00461 enhanced CRC progress by miR-323b-3p/NFIB axis.
Conclusion
In conclusion, our study first demonstrated that LINC00461 as an oncogenic promoted the proliferation, migration and invasion, and inhibited apoptosis of CRC cells by regulating the expression of miR-323b-3p, NFIB, p21 and CDK2. Our study may provide a reference for the diagnosis and treatment of CRC.
Acknowledgments
We kindly acknowledge The Third Hospital of Hebei Medical University who sponsored this research.
Author Contributions
Yulong Liang, Hairong Yu and Jianshuang Chen conceived of the study and designed experiments. Hairong Yu, Jianguo Ma and Jianshuang Chen carried out the experiments. Yang Yang and Jianjing Liang provided some reagents and carried out some experiments. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi:10.3322/CA.2007.0010
2. Xu L, Li X, Cai M, et al. Increased expression of Solute carrier family 12 member 5 via gene amplification contributes to tumour progression and metastasis and associates with poor survival in colorectal cancer. Gut. 2016;65(4):635–646. doi:10.1136/gutjnl-2014-308257
3. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi:10.3322/caac.21395
4. Lombardi L, Morelli F, Cinieri S, et al. Adjuvant colon cancer chemotherapy: where we are and where we’ll go. Cancer Treat Rev. 2010;36(Suppl 3):S34–S41. doi:10.1016/S0305-7372(10)70018-9
5. Alonso-Espinaco V, Cuatrecasas M, Alonso V, et al. RAC1b overexpression correlates with poor prognosis in KRAS/BRAF WT metastatic colorectal cancer patients treated with first-line FOLFOX/XELOX chemotherapy. Eur J Cancer. 2014;50(11):1973–1981. doi:10.1016/j.ejca.2014.04.019
6. Sugiyama M, Oki E, Nakaji Y, et al. High expression of the Notch ligand Jagged-1 is associated with poor prognosis after surgery for colorectal cancer. Cancer Sci. 2016;107(11):1705–1716. doi:10.1111/cas.2016.107.issue-11
7. Sun X, Suo J, Yan J. Immunotherapy in human colorectal cancer: challenges and prospective. World J Gastroenterol. 2016;22(28):6362–6372. doi:10.3748/wjg.v22.i28.6362
8. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi:10.1038/nature11233
9. Li Z, Yu X, Shen J. Long non-coding RNAs: emerging players in osteosarcoma. Tumour Biol. 2016;37(3):2811–2816. doi:10.1007/s13277-015-4749-4
10. Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116(4):751–762. doi:10.1161/CIRCRESAHA.116.303549
11. Yang Y, Ren M, Song C, et al. LINC00461, a long non-coding RNA, is important for the proliferation and migration of glioma cells. Oncotarget. 2017;8(48):84123–84139. doi:10.18632/oncotarget.20340
12. Ji D, Wang Y, Li H, Sun B, Luo X. Long non-coding RNA LINC00461/miR-149-5p/LRIG2 axis regulates hepatocellular carcinoma progression. Biochem Biophys Res Commun. 2019;512:176–181. doi:10.1016/j.bbrc.2019.03.049
13. Dong L, Qian J, Chen F, Fan Y, Long J. LINC00461 promotes cell migration and invasion in breast cancer through miR-30a-5p/integrin beta3 axis. J Cell Biochem. 2019;120(4):4851–4862. doi:10.1002/jcb.27435
14. Mirabello L, Koster R, Moriarity BS, et al. A genome-wide scan identifies variants in NFIB associated with metastasis in patients with osteosarcoma. Cancer Discov. 2015;5(9):920–931. doi:10.1158/2159-8290.CD-15-0125
15. Liu RZ, Vo TM, Jain S, et al. NFIB promotes cell survival by directly suppressing p21 transcription in TP53-mutated triple-negative breast cancer. J Pathol. 2019;247(2):186–198. doi:10.1002/path.5182
16. Reyes J, Chen JY, Stewart-Ornstein J, Karhohs KW, Mock CS, Lahav G. Fluctuations in p53 signaling allow escape from cell-cycle arrest. Mol Cell. 2018;71(4):581–591 e585. doi:10.1016/j.molcel.2018.06.031
17. Emmrich S, Streltsov A, Schmidt F, Thangapandi VR, Reinhardt D, Klusmann JH. LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer. 2014;13:171. doi:10.1186/1476-4598-13-171
18. Zhang J, Li Z, Liu L, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67(1):171–187. doi:10.1002/hep.29405
19. Marin-Bejar O, Mas AM, Gonzalez J, et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017;18(1):202. doi:10.1186/s13059-017-1331-y
20. Wang WT, Ye H, Wei PP, et al. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9(1):117. doi:10.1186/s13045-016-0348-0
21. Wang K, Long B, Zhou LY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi:10.1038/ncomms4596
22. Gao XR, Huang H, Kim H. Genome-wide association analyses identify 139 loci associated with macular thickness in the UK Biobank cohort. Hum Mol Genet. 2019;28(7):1162–1172. doi:10.1093/hmg/ddy422
23. Deng M, Yuan H, Liu S, Hu Z, Xiao H. Exosome-transmitted LINC00461 promotes multiple myeloma cell proliferation and suppresses apoptosis by modulating microRNA/BCL-2 expression. Cytotherapy. 2019;21(1):96–106. doi:10.1016/j.jcyt.2018.10.006
24. Xie L, Yao Z, Zhang Y, et al. Deep RNA sequencing reveals the dynamic regulation of miRNA, lncRNAs, and mRNAs in osteosarcoma tumorigenesis and pulmonary metastasis. Cell Death Dis. 2018;9(7):772. doi:10.1038/s41419-018-0813-5
25. Gonzalez-Vallinas M, Rodriguez-Paredes M, Albrecht M, et al. Epigenetically regulated chromosome 14q32 miRNA cluster induces metastasis and predicts poor prognosis in lung adenocarcinoma patients. Mol Cancer Res. 2018;16(3):390–402. doi:10.1158/1541-7786.MCR-17-0334
26. Navarro-Quiroz E, Pacheco-Lugo L, Navarro-Quiroz R, et al. Profiling analysis of circulating microRNA in peripheral blood of patients with class IV lupus nephritis. PLoS One. 2017;12(11):e0187973. doi:10.1371/journal.pone.0187973
27. Meng Y, Quan L, Liu A. Identification of key microRNAs associated with diffuse large B-cell lymphoma by analyzing serum microRNA expressions. Gene. 2018;642:205–211. doi:10.1016/j.gene.2017.11.022
28. Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249(1–2):31–45. doi:10.1016/S0378-1119(00)00140-2
29. Campbell TM, Castro MAA, de Oliveira KG, Ponder BAJ, Meyer KB. ERalpha binding by transcription factors NFIB and YBX1 enables FGFR2 signaling to modulate estrogen responsiveness in breast cancer. Cancer Res. 2018;78(2):410–421. doi:10.1158/0008-5472.CAN-17-1153
30. Stringer BW, Bunt J, Day BW, et al. Nuclear factor one B (NFIB) encodes a subtype-specific tumour suppressor in glioblastoma. Oncotarget. 2016;7(20):29306–29320. doi:10.18632/oncotarget.8720
31. Brun M, Jain S, Monckton EA, Godbout R. Nuclear factor I represses the notch effector HEY1 in glioblastoma. Neoplasia. 2018;20(10):1023–1037. doi:10.1016/j.neo.2018.08.007
32. Perez-Casellas LA, Wang X, Howard KD, Rehage MW, Strong DD, Linkhart TA. Nuclear factor I transcription factors regulate IGF binding protein 5 gene transcription in human osteoblasts. Biochim Biophys Acta. 2009;1789(2):78–87. doi:10.1016/j.bbagrm.2008.08.013
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.