Back to Journals » Cancer Management and Research » Volume 11
lncRNA LINC00460 promoted colorectal cancer cells metastasis via miR-939-5p sponging
Authors Zhang Y, Liu XC , Li Q, Zhang Y
Received 27 October 2018
Accepted for publication 17 December 2018
Published 22 February 2019 Volume 2019:11 Pages 1779—1789
DOI https://doi.org/10.2147/CMAR.S192452
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Yueyan Zhang,1* Xingchi Liu,2* Qiang Li,1* Yong Zhang1
1Department of Pathology, Liaoning Cancer Hospital and Institute, Cancer Hospital of China Medical University, Shenyang, People’s Republic of China; 2Department of Thoracic Surgery, General Hospital of Shenyang Military Command, Shenyang, People’s Republic of China
*These authors contributed equally to this work
Background: lncRNAs are widely involved in multiple malignancies including colorectal cancer (CRC). The expression and function of long intergenic non-protein coding RNA 460 (LINC00460) in CRC remains obscure.
Methods: In the present study, quantitative real-time PCR assays were applied to detect the expression changes of LINC00460 and microRNA-939-5p (miR-939-5p) in CRC tissue specimens and cell lines. Western blot assays were used to measure the changes of LIMK2. Bioinformatics analysis, luciferase assays, and RNA pull-down assays were applied to determine the targeting binding effect between LINC00460 and miR-939-5p as well as LIMK2 and miR-939-5p. Transwell assays were used to evaluate the metastatic ability changes of CRC line HT29 and LOVO cells.
Results: We found that LINC00460 was upregulated and closely correlated to clinicopathological features and poor prognosis of patients with CRC. Functionally, we elucidated that LINC00460 promoted metastasis in CRC cell lines HT29 and LOVO. Further, we showed that LIMK2 was a downstream effector in the LINC00460-induced promotion of metastasis in CRC cells HT29 and LOVO. Through online bioinformatics analysis, LINC00460 and LIMK2 were demonstrated to share similar microRNA response elements for miR-939-5p. Then, LINC00460 and LIMK2 were verified to be the targets of miR-939-5p via a luciferase assay and an RNA pull-down assay. Also, miR-939-5p was showed to suppress metastasis by targeting of LIMK2. Lastly, we revealed that LINC00460 promoted LIMK2-mediated metastasis via miR-939-5p sponging in CRC cells HT29 and LOVO.
Conclusion: The findings of this study showed that LINC00460 works as an oncogene in CRC and promoted CRC cell metastasis via regulation of miR-939-3p/LIMK2 axial. The present study might provide a new target in treating CRC.
Keywords: LINC00460, miR-939-5p, ceRNA, metastasis, colorectal cancer
Introduction
As one of the leading causes of cancer mortality, colorectal cancer (CRC) ranks as the third most common malignancies worldwide.1 It is reported that about 50%–60% of patients with CRC die from distant metastasis, especially liver metastasis.2 Most CRC patients with liver metastasis have a short 5-year survival rate of <10%, because of their non-adaptation to surgical treatment.3 Therefore, it is of great urgency to identify new molecules and the underlying mechanisms associated with metastasis development.
lncRNAs are a class of novel identified transcribed RNAs with >200 nucleotides in length.4 Recent evidences have revealed that lncRNAs frequently participate in various diseases, particularly in malignancies.5–7 As a member of lncRNAs, long intergenic non-protein coding RNA 460 (LINC00460) is located on chromosome 13q33.2 and is involved in several cancers. Liang et al showed that LINC00460 was overexpressed and promoted carcinogenesis in esophageal squamous cell carcinoma.8 Xing et al reported that LINC00460 was upregulated in meningioma tissues and malignant meningioma cell lines. They also found that LINC00460 promoted meningioma progression and metastasis via targeting of miR-539/MMP-9.9 To date, related research about the expression and function of LINC00460 in CRC metastasis and its specific working mechanism remains ambiguous.
Accumulative evidence has demonstrated that lncRNAs could indirectly regulate their downstream genes by acting as ceRNA, which was first proposed by Leonardo Salmena. ceRNA theory hypothesizes that RNA transcriptions could communicate through a new “language” mediated by microRNA response elements (MREs).10 Wang et al reported that lncRNA differentiation antagonizing non-protein coding RNA promoted heat shock protein 27-mediated proliferation and metastasis via acting as a ceRNA of miR-577 in CRC.11 Huang et al showed that lncRNA cancer susceptibility candidate 2 functioned as a ceRNA by miR-18a sponging in CRC.12 Till date, whether LINC00460 could function as ceRNA in CRC metastasis remains to be explored.
In the present study, we addressed the expression and function of LINC00460 in CRC. We elucidated that LINC00460 works as an oncogene to promote LIMK2-mediated metastasis through acting as a ceRNA of microRNA-939-5p (miR-939-5p) in CRC.
Patients and methods
Patients and tissue samples
Seventy-four cases of CRC tissue specimens and paired para-tumor specimens were collected during tumorectomy in Liaoning Cancer Hospital & Institute between April 2008 and April 2013. Written informed consent was obtained from patients whose tissues were used in the present study. The present study was conducted in accordance with the Declaration of Helsinki and was granted approval by the Institute Research Medical Ethics Committee of Liaoning Cancer Hospital & Institute. All 74 cases were diagnosed according to a definite pathological diagnosis, and the clinical stage of these patients was determined according to the TNM classification of the International Union Against Cancer.
Cell culture
A normal human colon epithelial cell line NCM460, and human colon cancer cell lines HT29, HCT116, SW480, and LOVO were purchased from the American Type Culture Collection and cultured in RPMI1640 medium (Thermo Fisher Scientific, Waltham, MA, USA). All media were supplemented with 10% (v/v) FBS (Sigma-Aldrich Co., St Louis, MO, USA), 100 IU/mL penicillin, and 100 mg/mL streptomycin (Baomanbio, Shanghai, People’s Republic of China). All cells were conditioned at 37°C in a humidified atmosphere containing 5% CO2.
Reverse transcription and quantitative real-time PCR (qRT-PCR)
The procedure was carried out as previously described.13 Total RNA from CRC tissue specimens and CRC cells were extracted by using a Trizol reagent (Thermo Fisher Scientific), respectively. cDNAs were synthesized by a Takara RNA PCR kit (Takara, Dalian, People’s Republic of China) according to the manufacturer’s protocol. PCR reactions containing SYBR Premix Ex Taq II (Takara) were performed according to the manufacturer’s instructions. GAPDH and U6 were used to normalize the expression levels of LINC00460/LIMK2 and miR-939-5p, individually. The relative gene expression was calculated using the 2−ΔΔCt method. Primer sequences were synthesized by RiboBio (Guangzhou, People’s Republic of China) (Table 1).
  | Table 1 Sequences of PCR primers used for RNA analysis |
Oligonucleotide and plasmid transfection
Effective siRNA oligonucleotides that targeted LINC00460 (siLINC00460-1 and siLINC00460-2) and LIMK2 (siLIMK2-1 and siLIMK2-2) and scramble control (siSCR) were chemically synthesized by RiboBio. miR-939-5p mimics and negative control (NC mimic), miR-939-5p inhibitor and negative control (NC inhibitor) were also purchased from RiboBio. Wild LINC00460 overexpression plasmids oeLINC00460/oeLINC00460-wt (containing a miR-939-5p response elements [MREs-939-5p]), mutant LINC00460 overexpression plasmids oeLINC00460-mut (containing a mutant MREs-939-5p), and LIMK2 overexpression plasmids (oeLIMK2) were purchased from GenePharma (Suzhou, People’s Republic of China). To obtain different expression levels of targeted gene cell models, we transfected all the oligonucleotides and plasmids into HT29 and LOVO cells by using a Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions, individually.
Transwell assay
The procedure was performed as previously described.14 In short, HT29 and LOVO cells were plated in the upper uncoated (for migration) and Matrigel-coated (for invasion) chambers (BD Biosciences, San Jose, CA, USA) with serum-free medium, respectively. Culture medium with 10% FBS was supplemented into the lower wells. All chambers were incubated for a further 24 hours. Twenty-four hours later, the non-migrated or non-invaded cells were wiped out. Then, the filters were fixed and stained by crystal violet staining. Five random fields were counted per chamber by using an inverted microscope (Olympus Corporation, Tokyo, Japan).
Western blot analysis
Total protein from cells was isolated by using RIPA lysis buffer (Sigma-Aldrich Co.) and was qualified by using a BCA detecting kit (Keygen, Nanjing, People’s Republic of China). Proteins samples were subjected to 10% SDS-PAGE and transferred onto a PVDF membrane, and then incubated with LIMK2 (dilution rates of 1:2,000; Abcam, Cambridge, UK) and GAPDH antibodies (dilution rates of 1:500; Abcam) at 4°C overnight, respectively. The next day, the membranes were incubated with secondary antibodies (dilution rates of 1:2,000; Abcam) at room temperature for 1 hour. Protein bands were detected on X-ray film using an enhanced chemiluminescence detection system.
In situ hybridization (ISH) of RNA
The procedure was carried out as previously described.11 Briefly, CRC tissue slices were permeabilized with 0.5% Triton X-100 and then incubated with special probes targeting LINC00460 (anti-LINC00460) and miR-939-5p (anti-miR-939-5p) (RiboBio) in a hybridization solution containing 1% blocking solution in a humid chamber at 37°C overnight. The following day, the slices were orderly washed with a solution of 0.1% Tween-20 in 4× sodium citrate buffer (SSC) for 5 minutes, a solution of 0.1% Tween-20 in 2× SSC for 5 minutes, and a solution of 0.1% Tween-20 in 1× SSC for 5 minutes at 42°C in the dark. Lastly, the slices were triply rinsed with 1× PBS for 5 minutes at room temperature and were counterstained by DAPI (Cell Signaling Technologies, Danvers, MA, USA). All slices were observed and photographed under a microscope (Leica Microsystems, Wetzlar, Germany).
Dual luciferase reporter assay
The procedure was carried out as previously described.11 Reporter plasmids of LINC00460 (wt-LINC00460-luc and mut-LINC00460-luc) and LIMK2 (wt-LIMK2-luc and mut-LIMK2-luc) were synthesized by GenePharma (Shanghai, People’s Republic of China). When HT29 and LOVO cells grew to 70% confluence, the synthesized reporter plasmids were co-transfected with miR-939-5p mimic or NC mimic by using Lipofectamine 2000 (Thermo Fisher Scientific). Forty-eight hours after transfection, fluorescence signal changes in each group were evaluated by a Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA) according to the manufacturer’s protocol.
RNA pull-down assay
The procedure was carried out as previously described.15 LINC00460-wt and LINC00460-mut as well as LIMK2-wt and LIMK2-mut were transcribed from vector pGEM®-T (Promega Corporation) and biotin-labeled with the Biotin RNA Labeling Mix (Hoffman-La Roche Ltd., Basel, Switzerland) and T7 RNA polymerase (Hoffman-La Roche Ltd.), treated with RNase-free DNase I (Hoffman-La Roche Ltd.), and purified with an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The biotinylated LINC00460 and LIMK2 probes were dissolved in binding and washing buffer and incubated with Dynabeads M-280 Streptavidin (Thermo Fisher Scientific) at 25°C for 10 minutes to generate probe-coated beads according to the manufacturer’s protocol. Then, cell lysates of HT29 and LOVO were incubated with the probe-coated beads, and the RNA complexes bound to these beads were eluted and extracted for qRT-PCR analysis to detect the relative expression of miR-939-5p.
Statistical analysis
All experiments were repeated in triplicate, and all data from three independent experiments were expressed as mean ± SD. GraphPad Prism V5.0 (GraphPad Software, Inc., La Jolla, CA, USA) software and SPSS 19.0 statistical software were used for statistical analysis. Correlation between LINC00460 and clinicopathological features of CRC patients was analyzed by using the Pearson’s chi-squared test or Mann–Whitney U test. Survival analysis was performed using the log-rank test in GraphPad Prism V5.0. Differences in two groups were analyzed with the Student’s t-test or one-way ANOVA. Differences were considered significant or very significant if P-value <0.05 or 0.01, respectively.
Results
LINC00460 was upregulated and correlated with poor prognosis in CRC
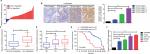
We first determined the expression of LINC00460 in 74 CRC tissue specimens and paired para-tumor tissue specimens by qRT-PCR. As the outcomes presented in Figure 1A, LINC00460 was upregulated in most (65/74, 87.84%) CRC tissue specimens. In addition, ISH analysis was used to measure the expression of LINC00460 in CRC tissue specimens with different staging. As the representative photographs illustrate in Figure 1B, gradually elevated LINC00460 levels was found with the advanced staging of CRC (P<0.001). Further, we found that upregulated LINC00460 was more commonly present in CRC tissue specimens with liver metastasis and lymph node metastasis (Figure 1C, D, P<0.001). According to the median expression of LINC00460 in the present research, we defined the patients with CRC as high LINC00460 group and low LINC00460 group. Even further, we analyzed the correlation between different levels of LINC00460 and clinicopathological features in patients with CRC. As the data show in Figure 1E and Table 2, high LINC00460 level was closely correlated with clinicopathological features, especially with shorter survival rate (Figure 1E, P<0.0001), clinical stage (P=0.038), metastasis (M) classification (P=0.005), nodal (N) classification (P=0.010), and liver metastasis (P=0.001) (Table 2). Lastly, we checked the expression of LINC00460 at cellular level. As the outcomes illustrate in Figure 1F, LINC00460 was significantly upregulated in four CRC cell lines (HT29, HCT116, SW480, and LOVO) as compared to a normal human colon epithelial cell line (NCM460) (P<0.001).
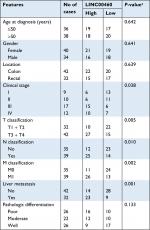  | Table 2 Correlation of LINC00460 expression and clinicopathological features in CRC Note: aP-value obtained from Pearson’s chi-squared test. Abbreviation: CRC, colorectal cancer. |
Knockdown of LINC00460 inhibited migration/invasion and decreased LIMK2 expression in HT29 and LOVO cells
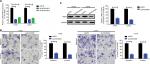
In the previous section, we found that upregulated LINC00460 was closely correlated with M classification, N classification, and liver metastasis. Therefore, we tried to explore the role that LINC00460 might play in metastasis of CRC. We first transfected LINC00460-specific siRNAs (siLINC00460-1 and siLINC00460-2) and siSCR into HT29 and LOVO cells to knockdown LINC00460. As shown in Figure 2A, siLINC00460-1 presented the silencing effect more efficiently and was selected for the following RNAi experiments (P<0.01). Then a transwell assay was performed to determine the changes in metastatic ability. Knockdown of LINC00460 significantly inhibited migration and invasion of HT29 and LOVO cells (Figure 2B).
LIMK2, a downstream effector of Rho/ROCK which contributes to Rho-induced reorganization of the actin cytoskeleton, was also reported to be a metastasis-related gene in CRC.16,17 Here, we speculated whether LINC00460 might regulate LIMK2 expression. Knockdown of LINC00460 remarkably decreased LIMK2 expression at protein level (P<0.01; Figure 2C).
LINC00460 promoted migration/invasion by upregulation of LIMK2 in HT29 and LOVO cells
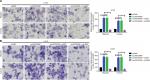
In the previous section, we showed that depression of LINC00460 inhibited LIMK2 expression and migration/invasion in CRC cells. We then attempted to clarify whether LIMK2 was a downstream effector in LINC00460-induced migration/invasion. Through a transwell assay, we illustrated that overexpression of LINC00460 promoted migration/invasion, and the facilitative effect was attenuated by depression of LIMK2 (co-transfection of oeLINC00460 + siLIMK2) in HT29 and LOVO cells. Meanwhile, the suppressive effect exhibited by siLINC00460 on migration/invasion was reversed by upregulation of LIMK2 (co-transfection of siLINC00460 + oeLIMK2) (Figure 3A, B, P<0.01). In brief, the findings of this analysis indicated that LINC00460 promoted migration/invasion through upregulation of LIMK2 in HT29 and LOVO cells.
miR-939-5p suppressed migration/invasion via directly targeting LIMK2 in HT29 and LOVO cells
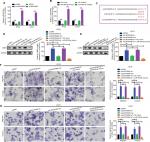
In the previous sections, we showed that LINC00460 promoted migration/invasion via upregulation of LIMK2 protein. We then tried to figure out the potential working mechanism between LINC00460 and LIMK2. We first showed that an up- and downregulation of LINC00460 did not affect LIMK2 mRNA expression significantly, and this phenomenon indicated that LINC00460 regulated LIMK2 expression at post-transcriptional level (Figure 4A). Recently, a most prevalent theory in nc RNA field is the ceRNA theory.10 Therefore, we suppose whether any miRNA might be involved in the regulative effect between LINC00460 and LIMK2. Online bioinformatics analysis (DIANA-LncBase and TargetScan) revealed that miR-939-5p might provide similar MREs for both LINC00460 and LIMK2 (Figure 4B). We then demonstrated the negative correlations between miR-939-5p and LINC00460 as well as miR-939-5p and LIMK2 (Figure 4C, D). Through a luciferase assay and an RNA pull-down assay, we verified the targeted binding effect between miR-939-5p and LINC00460 as well as miR-939-5p and LIMK2 (Figure 4E–H). Functionally, we confirmed that an elevation of miR-939-5p suppressed migration/invasion. But the suppressive effect could be reversed by a wild LIMK2 overexpression plasmid (oeLIMK2-wt) which contained a wild miR-939-5p response element (MREs-939-5p) but cannot be done by a mutant LIMK2 overexpression plasmid whose MREs-939-5p was mutated (oeLIMK2-mut) (Figure 4I–K, P<0.01). Overall, the outcomes of this investigation indicated that miR-939-5p suppressed migration/invasion via direct targeting of LIMK2.
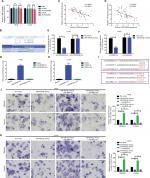  | Figure 4 miR-939-5p suppressed migration/invasion via directly targeting LIMK2 in HT29 and LOVO cells. Notes: (A) Up- and downregulation of LINC00460 did not affect LIMK2 mRNA expression as measured by a qRT-PCR assay. nsP>0.05 as normalizing and comparing with the mock group. (B) LINC00460 and LIMK2 shared a similar MRE for miR-939-5p as predicted by DIANA-LncBase (http://carolina.imis.athena-innovation.gr) and Targetscan (http://www.targetscan.org/vert_71). (C, D) Negative correlations between miR-939-5p and LIMK2 (C, P<0.0001) as well as miR-939-5p and LINC00460 (D, P=0.0001) were qualified by a Pearson’s correlation analysis. (E, F) Co-transfection of wt-LINC00460-luc (E) / wt-LIMK2-luc (F), and miR-939-5p mimic led to a significant weakening of luminescence as compared with mut-LINC00460-luc (E) / mut-LIMK2-luc (F), and miR-939-5p mimic co-transfection group, as determined by a luciferase assay. **P<0.01 and ns means non-significant as normalizing and comparing with NC mimic group. (G, H) Cell lysates were harvested from HT29 and LOVO cells and were incubated with biotin-labeled LINC00460-wt/LIMK2-wt and LINC00460-mut/LIMK2-mut individually. Subsequently, a qRT-PCR assay was performed to measure the pulled-down miR-939-5p in each group. As mentioned previously, it was LINC00460-wt/LIMK2-wt rather than LINC00460-mut/LIMK2-mut that could pull down miR-939-5p. **P<0.01 as normalizing and comparing to Beads group. (I) Diagram of the constructed LINC00460 or LIMK2 reporter plasmid containing a wild or a mutant MREs-939-5p. (J, K) Upregulation of miR-939-5p suppressed HT29 (J) and LOVO (K) cell metastasis, while the inhibitory effect was reversed by a wild LIMK2 overexpressing plasmid (oeLIMK2-wt) but not by a mutant one (oeLIMK2-mut). nsP>0.05; **P<0.01. Magnification, 4×; scale bars, 500 μm. Abbreviations: ns, non-significant; wt, wild type; mut, mutation; qRT-PCR, quantitative real-time PCR; siSCR, scramble control; NC, negative control. |
LINC00460 promoted migration/invasion through miR-939-5p sponging in HT29 and LOVO cells
In this section, we tried to elucidate whether the potential ceRNA mechanisms among LINC00460, miR-939-5p, and LIMK2 do exist. Firstly, we demonstrated the reciprocal inhibition effect between LINC00460 and miR-939-5p (Figure 5A, B). Secondly, we showed that a wild LINC00460 overexpression plasmid (oeLINC00460-wt) containing a MREs-939-5p promoted LIMK2 protein expression. When the theoretical MREs-939-5p was mutated (oeLINC00460-mut), the facilitative effect was dismissed. More convincingly, we illustrated that the facilitative effect of oeLINC00460-wt on LIMK2 protein could be attenuated by the miR-939-5p mimic (Figure 5C–E). In addition, we found that oeLINC00460-wt but not oeLINC00460-mut promoted migration/invasion in HT29 and LOVO cells, and the facilitative effect was reversed by the miR-939-5p mimic (Figure 5F, G).
Discussion
Increasing evidence has proven that ncRNAs play various roles in diverse cancer-related biological abnormal behaviors of a cell, including proliferation, apoptosis, angiogenesis, and metastasis.14,18,19 As a newly identified lncRNA, LINC00460 is located on chromosome 13q33.2 and contains three exon counts. Presently, related studies about LINC00460 are few. Yue et al found that LINC00460 was upregulated in non-small-cell lung cancer (NSCLC), and that depression of LINC00460 could inhibit the proliferation, invasion, and migration of NSCLC cells.20 In a study of nasopharyngeal carcinoma (NPC), Kong et al revealed that LINC00460 contributed to the progression of NPC through regulating miR-149-5p/IL6 signal pathway.21 In the present study, we showed that LINC00460 was upregulated in CRC tissue specimens and CRC cell lines. We also found that the upregulation of LINC00460 was correlated with CRC patients’ clinicopathological features, especially with M classification (P=0.005), N classification (P=0.010), and liver metastasis (P=0.001). Therefore, we concentrated on the role of LINC00460 in CRC cell metastasis. We illustrated that the silencing of LINC00460 inhibited CRC cell migration and invasion via a transwell assay.
MicroRNAs, 22–25 nucleotides, are another type of well-known ncRNAs. As a member of the miRNA families, miR-939-5p (also named as miR-939) was extensively involved in various malignant tumors including ovarian cancer, NSCLC, gastric cancer, breast cancer, and CRC.22–26 In our research, the expression of miR-939-5p was shown to correlate with LINC00460 and LIMK2 negatively; LIMK2 was revealed to be a direct downstream gene of miR-939-5p. miR-939-5p suppressed HT29 and LOVO cell metastasis via direct targeting of LIMK2. Moreover, we found that miR-939-5p could bind to LINC00460 and LIMK2 via similar miR-939-5p response elements (MREs-939-5p), and these findings indicated that miR-939-5p might work as a bridge connecting LINC00460 and LIMK2.
LIMK2, which belongs to the LIM kinase (LIMK) family, is a type of serine/threonine protein kinase with structure composed of two LIM motifs at the amino-terminus and an unusual protein kinase domain at the carboxy-terminus.27 As a downstream gene of Rho/ROCK pathway, activated LIMK2 could phosphorylate cofilin – a potent regulator of actin filament dynamics – and involve in ROCK-induced metastatic phenotype.28 Wang et al found that bone morphogenetic protein receptor 2 promoted invasion and metastasis via regulation of RhoA/ROCK/LIMK2 pathway in osteosarcoma.29 Aggelou et al reported that LIMK2 was overexpressed in CRC and was associated with CRC tumor progression parameters.30 In the present study, we found that a knockdown of LINC00460 inhibited LIMK2 protein expression. Also, we showed that the silencing of LIMK2 significantly attenuated LINC00460-induced metastasis, and this finding indicated that LIMK2 was a downstream effector in LINC00460-induced metastasis in CRC. Through a qRT-PCR assay, we revealed that LINC00460 did not affect LIMK2 mRNA expression and the phenomenon indicated that LINC00460 might regulate LIMK2 at a post-transcriptional level. To verify whether the ceRNA working mechanism does exist among LINC00460, LIMK2, and miR-939-5p, we first determined the relationship between miR-939-5p and LINC00460. A reciprocal inhibitive effect was found between miR-939-5p and LINC00460. We then demonstrated that only oeLINC00460-wt could promote metastasis and LIMK2 protein expression. When the theoretical MREs-939-5p was mutated, the facilitative effect was dismissed. More convincingly, we found that miR-939-5p mimic could significantly reverse the promotive effect of oeLINC00460-wt on CRC cell metastasis and LIMK2 protein expression. Taking all these events into account, the outcomes strongly indicated that LINC00460, working as an oncogene, promoted metastasis by upregulating LIMK2 via acting as a ceRNA of miR-939-5p in CRC (Figure 6).
Conclusion
The findings of this study showed that LINC00460 work as an oncogene in CRC and promoted CRC cells metastasis via regulating of miR-939-3p/LIMK2 axial. The present study might provide a new target in treating of CRC.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. | ||
Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. | ||
Landreau P, Drouillard A, Launoy G, et al. Incidence and survival in late liver metastases of colorectal cancer. J Gastroenterol Hepatol. 2015;30(1):82–85. | ||
Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222. | ||
Fanale D, Barraco N, Listì A, Bazan V, Russo A. Non-coding RNAs functioning in colorectal cancer stem cells. Adv Exp Med Biol. 2016;937:93–108. | ||
Khorshidi A, Dhaliwal P, Yang BB. Noncoding RNAs in tumor angiogenesis. Adv Exp Med Biol. 2016;927:217–241. | ||
Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592(17):2884–2900. | ||
Liang Y, Wu Y, Chen X, et al. A novel long noncoding RNA linc00460 up-regulated by CBP/p300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017;37(5):BSR20171019. | ||
Xing H, Wang S, Li Q, Ma Y, Sun P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed Pharmacother. 2018;105:677–682. | ||
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. | ||
Wang Y, Lu Z, Wang N, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50(5):57. | ||
Huang G, Wu X, Li S, Xu X, Zhu H, Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci Rep. 2016;6(1):26524. | ||
Wang Y, Zhang Y, Yang T, et al. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8(35):59417–59434. | ||
Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89. | ||
Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8(1):289. | ||
Lourenço FC, Munro J, Brown J, et al. Reduced LIMK2 expression in colorectal cancer reflects its role in limiting stem cell proliferation. Gut. 2014;63(3):480–493. | ||
Zhang Y, Li A, Shi J, et al. Imbalanced LIMK1 and LIMK2 expression leads to human colorectal cancer progression and metastasis via promoting β-catenin nuclear translocation. Cell Death Dis. 2018;9(7):749. | ||
Cheng Z, Li Z, Ma K, et al. Long non-coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J Cancer. 2017;8(19):4106–4116. | ||
Sun Y, Qin B. Long noncoding RNA MALAT1 regulates HDAC4-mediated proliferation and apoptosis via decoying of miR-140-5p in osteosarcoma cells. Cancer Med. 2018;7(9):4584–4597. | ||
Yue QY, Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(4):1003–1010. | ||
Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. lncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. | ||
Cui C, Zhai D, Cai L, Duan Q, Xie L, Yu J. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939-mediated transcriptional repression of Bcl-xL. Cancer Res Treat. 2018;50(3):992–1008. | ||
Di Modica M, Regondi V, Sandri M, et al. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017;384:94–100. | ||
Ma R, Wang C, Wang J, Wang D, Xu J. miRNA-mRNA interaction network in non-small cell lung cancer. Interdiscip Sci. 2016;8(3):209–219. | ||
Ying X, Li-ya Q, Feng Z, Yin W, Ji-Hong L. miR-939 promotes the proliferation of human ovarian cancer cells by repressing Apc2 expression. Biomed Pharmacother. 2015;71:64–69. | ||
Zhang JX, Xu Y, Gao Y, et al. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol Cancer. 2017;16(1):18. | ||
Hiraoka J, Okano I, Higuchi O, Yang N, Mizuno K. Self-association of LIM-kinase 1 mediated by the interaction between an N-terminal LIM domain and a C-terminal kinase domain. FEBS Lett. 1996;399(1-2):117–121. | ||
Suyama E, Wadhwa R, Kawasaki H, et al. LIM kinase-2 targeting as a possible anti-metastasis therapy. J Gene Med. 2004;6(3):357–363. | ||
Wang S, Ren T, Jiao G, et al. BMPR2 promotes invasion and metastasis via the RhoA-ROCK-LIMK2 pathway in human osteosarcoma cells. Oncotarget. 2017;8(35):58625–58641. | ||
Aggelou H, Chadla P, Nikou S, et al. LIMK/cofilin pathway and slingshot are implicated in human colorectal cancer progression and chemoresistance. Virchows Arch. 2018;472(5):727–737. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.