Back to Journals » Cancer Management and Research » Volume 12
lncRNA LEF1-AS1 Promotes Proliferation and Induces Apoptosis of Non-Small-Cell Lung Cancer Cells by Regulating miR-221/PTEN Signaling
Authors Xiang C, Zhang Y, Zhang Y, Liu C, Hou Y, Zhang Y
Received 17 January 2020
Accepted for publication 31 March 2020
Published 25 May 2020 Volume 2020:12 Pages 3845—3850
DOI https://doi.org/10.2147/CMAR.S246422
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Chen Xiang,1 Yuanli Zhang,2 Yajing Zhang,1 Ci Liu,1 Yuehong Hou,1 Yan Zhang1
1Department of Oncology IV, First Hospital of Shijiazhuang, Shijiazhuang City, Hebei Province 050000, People’s Republic of China; 2Department of Cardiology Ⅱ, First Hospital of Shijiazhuang, Shijiazhuang City, Hebei Province 050000, People’s Republic of China
Correspondence: Yan Zhang
Department of Oncology IV, First Hospital of Shijiazhuang, No. 36 Fanxi Road, Shijiazhuang City, Hebei Province 050000, People’s Republic of China
Tel +86 133 1597 8336
Email [email protected]
Introduction: LEF1-AS1 is a characterized oncogenic lncRNA in oral cancer. Analysis of TCGA dataset revealed the upregulation of LEF1-AS1 in non-small-cell lung cancer (NSCLC). This study was therefore carried out to investigate the involvement of LEF1-AS1 in NSCLC.
Methods: A total of 62 NSCLC patients were included to collect paired cancer and non-tumor tissues. RT-qPCR was performed to measure levels of LEF1-AS1 and miR-221 expression. Transient transfections were performed to explore the interactions between LEF1-AS1, miR-221 and PTEN. Cell proliferation and apoptosis were analyzed by cell proliferation assay and cell apoptosis assay, respectively.
Results: We found that LEF1-AS1 was upregulated in NSCLC patients. In addition, expression of LEF1-AS1 was negatively correlated with the expression of PTEN but positively correlated with the expression of miR-221 in NSCLC tissue samples. In NSCLC cells, overexpression of LEF1-AS1 led to downregulated expression of PTEN but upregulated expression of miR-221, which can directly target PTEN. Overexpression of LEF1-AS1 and miR-221 promoted cancer cell proliferation and inhibited apoptosis. PTEN played an opposite role and reduced the effects of overexpressing LEF1-AS1 and miR-221.
Conclusion: LEF1-AS1 may promote the proliferation and induce apoptosis of NSCLC cells by regulating miR-221/PTEN signaling.
Keywords: non-small-cell lung cancer, LEF1-AS1, miR-221, PTEN
Background
Lung cancer is the most common type of malignancy in males and the third most common type of malignancy in females.1 In 2018 only, lung cancer caused 1,761,007 deaths, which accounted for 18.4% of all cancer-related deaths, and it affected 2,093,876 new cases, accounting for 11.6% of all new cancer cases in the same year.2 The most common type of lung cancer is non-small-cell lung cancer (NSCLC), which accounts for more than 85% of all lung cancer cases.3 Smoking is the major risk factor for NSCLC, while NSCLC also affects never-smokers,4 indicating the complicated pathogenesis of this disease. In addition, NSCLC can be divided into two major subtypes, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), which have different pathogenesis.5,6 The treatment of NSLC is also complicated.
Molecular signaling pathways are crucial players in the pathogenesis of NSCLC.7 Analysis of the functions of the molecular pathways in the occurrence, development and progression of NSCLC provides novel insights into the development of targeted therapies.8 Long non-coding RNAs (lncRNAs) in recent years have been shown to have critical functions in cancer biology.9 LncRNAs do not encode proteins but regulate protein synthesis at different levels to regulate cancer-related gene expression.10 Therefore, functional characterization of lncRNAs in cancer biology will provide novel targets for anti-cancer therapy. However, the roles of most lncRNAs in cancer biology are still unknown. LEF1-AS1 is a recently characterized oncogenic lncRNA in different types of cancer including lung cancer.11–13 We analyzed the TCGA database and observed the upregulation of LEF1-AS1 in both LUAD and LUSC. In addition, our preliminary RNA-seq data revealed positive correlation between LEF1-AS1 and miR-221, which plays oncogenic roles mainly by targeting tumor suppressor PTEN.14 This study was therefore carried out to investigate the interactions among LEF1-AS1, miR-221 and PTEN in NSCLC.
Methods
NSCLC Patients
This study enrolled 62 cases of NSCLC patients (48 males and 14 females; age range 48 to 67 years old; mean age 55.3 ± 5.0 years old) at the First Hospital of Shijiazhuang between March 2017 and March 2019. This study was approved by the Ethics Committee of this hospital. All the NSCLC patients were diagnosed for the first time and no other clinical disorders were observed. No therapy was initiated before the enrollment. The 62 NSCLC patients included 34 cases of LUAD and 28 cases of LUSC. All patients signed the informed consent.
Specimens and Cell Lines
Before therapy, all patients were subjected to fine needle aspiration biopsy to collect paired cancer and adjacent (within 3cm around tumors) non-cancer tissue samples from each patient. All tissue samples were confirmed by histopathological exam and the fresh samples were stored in liquid nitrogen before use. NSCLC cell lines NCI-H1993 (H1993, LUAD) and NCI-H1581 (H2170, LUSC) were used in this study. Cells of both cell lines were obtained from ATCC (USA). Cells were cultivated in a mixture composed of 90% RPMI-1640 medium and 10% FBS. Cell culture conditions were 37°C, 5% CO2 and 95% humidity. Cells were collected at about 85% confluence to perform the following experiments.
Transient Cell Transfection
With pcDNA3.1 vector (Invitrogen) as the backbone, expression vectors of LEF1-AS1 and PTEN were constructed. MiR-221 mimic and miRNA negative control (NC) were purchased from Sigma-Aldrich. H1993 and H2170 cells were transfected with 10 nM vector or 45 nM miRNA using Lipofectamine 2000 reagent (Invitrogen). Untransfected cells were used as control (C) cells. Cells transfected with miRNA NC- or empty vector-transfected were used as NC cells. Cells were collected at 48 h after transfection.
RNA Preparations and RT-qPCR
Tissue samples and cells were subjected to total RNAs extraction using Ribozol reagent (Invitrogen). All RNA samples were subjected to genomic DNA removal using gDNA eraser (Takara). RNA precipitation and washing steps were performed using 85% ethanol to harvest miRNAs. The QuantiNova Reverse Transcription Kit (QIAGEN) was used to reverse transcribe total RNAs into cDNAs. The qPCR reactions were performed using the PowerUp SYBR™ Green Master Mix (Thermo Fisher Scientific) to measure the expression levels of LEF1-AS1 and PTEN. GAPDH was used as endogenous control for LEF1-AS1 and PTEN mRNA. The 18S rRNA was also used as the endogenous control of LEF1-AS1. Similar results were obtained. The expression levels of mature miR-221 were measured using the All-in-One™ miRNA qRT-PCR Reagent Kit (Genecopoeia) with U6 as endogenous control. All PCR reactions were performed in triplicate manner and the 2−ΔΔCT method was used for data normalization.
Western Blot
Proteins were isolated from H1993 and H2170 cells using RIPA buffer (Invitrogen). Protein concentration was measured using BCA assay (Invitrogen). Protein denaturation was performed in boiling water for 8 min. Protein separation was performed using SDS-PAGE gel (8%), and the separated proteins were transferred to PVDF membranes. Membrane blocking was performed using PBS containing 5% non-fat milk at room temperature for 2 h. After that, PTEN (ab31392, Abcam) and GAPDH (ab9485, Abcam) rabbit primary antibodies were used to incubate with the membranes at 4 °C for 14 h, followed by incubation with HRP Goat Anti-Rabbit secondary antibody (IgG, ab97051, Abcam) at room temperature for another 2 h. Signals were produced using ECL (Sigma-Aldrich) and data were quantified using Quantity One software (Bio-Rad).
Cell Counting Kit-8 (CCK-8) Analysis
The proliferation of H1993 and H2170 cells after transfection was assessed by CCK-8 assay (Dojindo). Each well of a 96-well plate was filled with 0.1 mL medium containing 5000 cells. Cells were cultivated at 37 °C and CCK-8 solution was added into each well to reach 10% final concentration at 4 h before the measurement of OD values. OD values were measured every 24 h for a total of 4 days.
Cell Apoptosis Analysis
H1993 and H2170 cells were subjected to cell apoptosis assay at 48 h post-transfection. Cells were treated with 0.25% trypsin digestion, followed by staining with propidium iodide and Annexin V-FITC (Dojindo, Japan) for 15 min in dark. Flow cytometry was performed to separate apoptotic cells.
Statistical Analysis
Mean ± SD values were used to express data from three independent replicates. Paired t-test was used to compare differences between paired tissues. ANOVA Tukey’s test was used to compare differences among multiple groups. Correlations were analyzed by linear regression. P < 0.05 was considered as statistically significant.
Results
LEF1-AS1 Was Upregulated in Both LUAD and LUSC Patients
The differential expression of LEF1-AS1 in NSCLC was first assessed by exploring the TCGA dataset. It was observed that expression levels of LEF1-AS1 were higher in both LUAD (0.32 vs.0.18) and LUSC (0.59 vs. 0.18) tissues than in non-tumor tissues. To further confirm the upregulation of LEF1-AS1 in NSCLC, expression levels of LEF1-AS1 in paired tissue samples from both 34 cases of LUAD and 28 cases of LUSC were measured by RT-qPCR. Compared to non-tumor tissues, the expression levels of LEF1-AS1 were significantly higher in cancer tissues in both LUAD (Figure 1A, p < 0.01) and LUSC (Figure 1B, p < 0.01) patients.
LEF1-AS1 Was Inversely Correlated with PTEN but Positively Correlated with miR-221
Expression levels of PTEN and miR-221 in cancer tissues were also measured by RT-qPCR. Linear regression was performed to analyze the correlation between the expression of LEF1-AS1 and PTEN or miR-221. It was observed that expression of LEF1-AS1 and PTEN were inversely correlated (Figure 2A), while expression of LEF1-AS1 and miR-221 were positively correlated (Figure 2B) across cancer tissue samples.
Overexpression of LEF1-AS1 Led to Downregulated PTEN but Upregulated miR-221 in NSCLC Cells
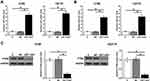
H1993 and H2170 cells were transfected with LEF1-AS1 expression vector, and the overexpression of LEF1-AS1 was confirmed by RT-qPCR at 48 h post-transfection (Figure 3A, p < 0.05). RT-qPCR and Western blot were performed to assess the effects of overexpression of LEF1-AS1 on the expression of miR-221 and PTEN, respectively. Compared to C and NC group, overexpression of LEF1-AS1 led to upregulated miR-221 (Figure 3B, p < 0.05). In contrast, overexpression of LEF1-AS1 resulted in downregulated PTEN in cells of both cell lines (Figure 3C, p < 0.05).
LEF1-AS1 Promoted NSCLC Proliferation and Inhibited Cell Apoptosis Through miR-221/PTEN Axis
Cell proliferation and apoptosis assays were performed to evaluate the effects of overexpressing LEF1-AS1, miR-221 and PTEN on the proliferation (Figure 4A) and apoptosis (Figure 4B) of NSCLC cells. It was observed that overexpression of LEF1-AS1 and miR-221 promoted cancer cell proliferation and inhibited apoptosis. PTEN played an opposite role and reduced the effects of overexpressing LEF1-AS1 and miR-221 (p < 0.05).
Discussion
In this study, the interactions among LEF1-AS1, miR-221 and PTEN were investigated. The results revealed that LEF1-AS1 was upregulated in NSCLC and it may upregulate miR-221 to downregulate PTEN, which resulted in inhibition of cancer cell apoptosis and promotion of cell proliferation.
The oncogenic function of LEF1-AS1 has been characterized in several types of cancer.11–13 For instance, LEF1-AS1 is upregulated in oral cancer and its knockdown led to inhibited cancer progression through the Hippo signaling pathway.11 LEF1-AS1 is also upregulated in glioblastoma and promotes tumor growth through the interactions with ERK and Akt/mTOR signaling.12 A recent study reported that overexpression of LEF1-AS1 in lung cancer regulated miR-544a/FOXP1 axis to promote cancer cell invasion and proliferation.13 However, lung cancer has several subtypes, for example, NSCLC has LUAD and LUSC two major subtypes.5,6 Therefore, the molecular pathogenesis of different subtypes of NSCLC should be studied separately. In this study, we showed that LEF1-AS1 was upregulated in both LUSC and LUAD. In addition, overexpression of LEF1-AS1 led to inhibited apoptosis and promoted proliferation of both LUAD and LUAS cells. Therefore, LEF1-AS1 may play similar roles in different subtypes of NSCLC.
A recent study reported that miR-221 played an oncogenic role in NSCLC by targeting TIMP2.15 In another study, miR-221 was reported to promote the progression of cutaneous squamous cell carcinoma by directly targeting PTEN.14 In the present study, the results demonstrated that miR-221 could also regulate apoptosis and proliferation of NSCLC cells by targeting PTEN. The key finding of the present study is that LEF1-AS1 may upregulate miR-221 to downregulate PTEN. However, the mechanism remains unclear. Our future studies will focus on elucidating the mediated regulation of miR-221 expression by LEF1-AS1.
In conclusion, LEF1-AS1 is upregulated in NSCLC and it can upregulate miR-221 to downregulate PTEN, thereby inhibiting cancer cell apoptosis and promoting cell proliferation.
Abbreviations
NSCLC, non-small-cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; lncRNAs, Long non-coding RNAs.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of First Hospital of Shijiazhuang. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients provided written informed consent prior to their inclusion within the study.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Natural Science Foundation of Hebei Province (H2019106033).
Disclosure
The authors declare that they have no competing interests.
References
1. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19.
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet. 2011;378(9804):1727–1740. doi:10.1016/S0140-6736(10)62101-0
4. Couraud S, Zalcman G, Milleron B, et al. Lung cancer in never smokers – a review. Eur J Cancer. 2012;48(9):1299–1311. doi:10.1016/j.ejca.2012.03.007
5. Chen M, Liu X, Du J, et al. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget. 2017;8(1):133–144. doi:10.18632/oncotarget.13346
6. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi:10.1038/nature25183
7. Kadara H, Scheet P, Wistuba II, et al. Early events in the molecular pathogenesis of lung cancer. Cancer Prev Res (Phila). 2016;9(7):518–527. doi:10.1158/1940-6207.CAPR-15-0400
8. Minna JD, Fong K, Zöchbauer-Müller S, et al. Molecular pathogenesis of lung cancer and potential translational applications. Cancer J. 2002;8:S41–S46.
9. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom Proteom Bioinf. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
10. Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi:10.1038/onc.2011.621
11. Zhang C, Bao C, Zhang X, et al. Knockdown of lncRNA LEF1-AS1 inhibited the progression of oral squamous cell carcinoma (OSCC) via Hippo signaling pathway. Cancer Biol Ther. 2019;20(9):1213–1222. doi:10.1080/15384047.2019.1599671
12. Wang J, Liu X, Yan C, et al. LEF1-AS1, a long-noncoding RNA, promotes malignancy in glioblastoma. Onco Targets Ther. 2017;10:4251–4260. doi:10.2147/OTT.S130365
13. Wang A, Zhao C, Gao Y, et al. LEF1-AS1 contributes to proliferation and invasion through regulating miR-544a/FOXP1 axis in lung cancer. Invest New Drugs. 2019;37(6):1127–1134. doi:10.1007/s10637-018-00721-z
14. Gong ZH, Zhou F, Shi C, et al. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol Biol Lett. 2019;24(1):9. doi:10.1186/s11658-018-0131-z
15. Yin Z, Xu M, Li P. miRNA-221 acts as an oncogenic role by directly targeting TIMP2 in non-small-cell lung carcinoma. Gene. 2017;620:46–53. doi:10.1016/j.gene.2017.04.007
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.