Back to Journals » Drug Design, Development and Therapy » Volume 14
LncRNA KCNQ1OT1 Sponges miR-206 to Ameliorate Neural Injury Induced by Anesthesia via Up-Regulating BDNF
Received 9 April 2020
Accepted for publication 23 September 2020
Published 9 November 2020 Volume 2020:14 Pages 4789—4800
DOI https://doi.org/10.2147/DDDT.S256319
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Qiongyu Guo
Yao Yao,* Xuesong Wang,* Jin Gao
Department of Anesthesiology, Xiangyang Central Hospital, Affiliated Hospital of Hubei College of Arts and Science, Xiangyang 441021, Hubei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jin Gao
Department of Anesthesiology, Xiangyang Central Hospital, Affiliated Hospital of Hubei College of Arts and Science, Jingzhou Street No. 39, Xiangcheng District, Xiangyang 441021, Hubei, People’s Republic of China
Tel +86 15897965817
Email [email protected]
Objective: Widely used in anesthesia, ketamine is reported to induce neurotoxicity in patients. This study aimed to investigate the molecular regulatory mechanism of long non-coding RNA (lncRNA) KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) in ameliorating ketamine-induced neural injury.
Materials and Methods: Sprague–Dawley rats were intraperitoneally injected with ketamine to induce neuronal injury. PC-12 cells treated with ketamine were used as the cell model. Ketamine-induced aberrant expression of KCNQ1OT1, miR-206 and brain-derived neurotrophic factor (BDNF) were examined by quantitative real-time polymerase chain reaction (qRT-PCR). The effects of KCNQ1OT1 and miR-206 on ketamine-induced neural injury in PC-12 cells were then examined by MTT and LDH assay. The regulatory relationships between KCNQ1OT1 and miR-206, and miR-206 and BDNF were detected by dual-luciferase reporter assay.
Results: Ketamine induced the apoptosis of neurons of the hippocampus in rats, and the apoptosis of PC-12 cells, accompanied by down-regulation of KCNQ1OT1 and BDNF expressions, and up-regulation of miR-206 expression. Overexpression of KCNQ1OT1 enhanced the resistance to apoptosis of PC-12 cells and significantly ameliorated ketamine-induced nerve injury, while transfection of miR-206 had opposite effects. Mechanistically, KCNQ1OT1 could target miR-206 and reduce its expression level, in turn indirectly increase the expression level of BDNF, and play a protective role in neural injury.
Conclusion: KCNQ1OT1/miR-206/BDNF axis is demonstrated to be an important regulatory mechanism in regulating ketamine-induced neural injury. Our study helps to clarify the mechanism by which ketamine exerts its toxicological effects and provides clues for the neuroprotection during anesthesia.
Keywords: ketamine, neural injury, lncRNA KCNQ1OT1, miR-206, BDNF
Introduction
Ketamine, a non-competing N-methyl-D-aspartate receptor antagonist, featuring with good clinical anesthetic effects on the circulatory and respiratory systems, it is widely used in pediatric wards. However, the potential adverse reactions caused by ketamine have attracted increasing attention in recent years. Both in vitro and in vivo studies suggest that ketamine has neurotoxicity, which may lead to neuronal apoptosis, thus inducing neurological impairment and affecting brain development and cognitive function.1–3 Therefore, it is particularly important to study the adverse reactions of ketamine, as well as to elucidate its pathophysiological mechanisms.4,5
Brain-derived neurotrophic factor (BDNF), which is generally expressed in central and peripheral nervous systems, is a member of neurotrophic factor family. BDNF stimulates and promotes the growth and differentiation of neurons mainly by binding to tyrosine kinase receptor tropomyosin-related kinase B (TrkB), regulating neuronal maturation, synapse formation and synaptic plasticity.6 It is reported that the BNDF released from neurons is inhibited by a high dose of ketamine, thus leading to nerve injury.7 It is also reported that baicalin has a neuroprotective effect against ketamine-induced neuronal apoptosis by upregulating BDNF expression.8 However, the molecular mechanism by which ketamine induces the down-regulation of BDNF expression remains unclear.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs that consist of more than 200 nucleotides (nt) without open reading frame or coding sequence, which participate in various biological processes, such as cell growth, differentiation and apoptosis.9–11 In the central nervous system, the role of lncRNAs have been gradually unveiled.12–15 For example, lncRNA NEAT1 plays a vital role in the process of bexarotene-induced functional recovery after traumatic brain injury,14 and lncRNA-MEG3 promotes subarachnoid hemorrhage-induced neuron injury by inhibiting PI3K/Akt pathway.15 LncRNA KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) is a novel lncRNA discovered in recent years, and its biological functions in some human diseases have been reported.16 For example, it is reported that KCNQ1OT1 is a critical regulator in preventing ischemia-reperfusion induced myocardial injury following acute myocardial infarction.17 However, the role of KCNQ1OT1 in ketamine-induced neuronal damage remains obscure.
MicroRNAs (miRNAs) regulate the gene expression at the post-transcriptional or translational level by binding to the 3ʹ untranslated region (3ʹUTR) of mRNA.18,19 Recently, accumulating studies indicate that miRNAs participate in regulating anesthesia-induced neurotoxicity. For example, down-regulation of miR-34c expression activates PKC-ERK pathway and reduced ketamine-induced hippocampal cell apoptosis.20 Furthermore, some studies unearth that up-regulation of miR-206 expression leads to nerve damage by negatively modulating BDNF expression.21,22 However, little is known concerning the specific mechanism of how miR-206 regulates ketamine-induced neurological damage.
In this study, we aimed to investigate the role of KCNQ1OT1 and miR-206 in ketamine-induced neuronal injury. It was demonstrated that KCNQ1OT1 expression in rat brain embryonic stem cell (ESC)-derived neurons were suppressed by ketamine. Gain-function and loss-function experiments revealed that KCNQ1OT1 could target miR-206 and indirectly increase BDNF expression, thereby ameliorating ketamine-induced nerve damage. This study provided a new theoretical basis for the rational use of anesthetics.
Materials and Methods
Animal Model
All of the animal experiments were ratified by the Institutional Animal Care and Use Committee of Xiangyang Central Hospital (Approval Number: 2017101305; Approval date: October 13, 2017). All procedures in animal experiments were performed in accordance with the Guide for Care and Use of Laboratory Animals from the National Health Research Institutes, and Recommendations on Animal Care from the Chinese Neuroscience Society. Male Sprague–Dawley (SD) rats (50-day-old) with body weights ranging from 150 g to 200 g were maintained with a suitable temperature and humidity, free access to food and water at 12 h/12 h light/dark cycle. Twenty rats were randomly divided into two groups: ketamine group (n=15) and control group (n=5). In the ketamine group, rats (n=5 in each group) were intraperitoneally injected with 7.5, 15 and 30 mg/kg ketamine (Sigma, Shanghai, China) respectively, once a day for 3 consecutive days; in control group, rats (n=5) were injected with normal saline at the same interval (Figure 1A). Twenty-four hours after the last administration, the rats were anesthetized and euthanized to collect hippocampus tissues, and these tissues were stored in a refrigerator at – 80 °C for subsequent experiments.23
Fifteen rats (n=5 in each group) received stereotaxic injection to investigate the role of KCNQ1OT1 in vivo. A stereotaxic frame (David Kopf, Tujunga, CA, USA) was used to perform stereotactic surgery for rats, and 2.5 μL Hamilton syringe (Hamilton Medical, Reno, NV, USA) and bilateral microsyringe pump controller were used to inject recombinant adeno-associated virus (rAAV) into the hippocampus of the rats. Injection coordinates were as below: front and back: + 2.1 mm; medial lateral: ± 1.5 mm; dorsal ventral: −1.8 mm. In the control group, ketamine was not used, and normal saline was injected into the hippocampus; in ketamine + vector group and ketamine + KCNQIOT1 group, 30 mg/kg ketamine was intraperitoneally injected into each rat, and rAAV was injected into the hippocampus. The total injection volume of rAAV solution (or normal saline) was 0.75 μL, and the injection rate was kept at 0.25 μL/min.
Cell Culture and Treatment
Neuronal cell line PC-12 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Grand Island, NY, USA) supplemented with 10% horse serum (Gibco, Grand Island, NY, USA) and 5 % fetal bovine serum (Gibco, Grand Island, NY, USA). These cells were cultured in a humidified incubator containing 5% CO2 at 37 °C, and the medium was replaced every 48 h. Cells were trypsinized with 0.25% trypsin and sub-cultured. Cells in the logarithmic phase were harvested for further experiments. To establish the injury models, the cells were treated with 25 μM, 50 μM and 100 μM ketamine and cultured for 24 h.
Transfection
Specific mimics, inhibitors and the corresponding negative controls (mimic NC and inhibitor NC) of miR-206 were synthesized by GenePharma (Shanghai, China). The entire coding sequence of KCNQ1OT1 was cloned into pcDNA3.1 plasmid (Sangon Biotech, Shanghai, China) for KCNQ1OT1 overexpression plasmid (pc-KCNQ1OT1). Short-hairpin RNAs (shRNAs) specific to KCNQ1OT1 (si-KCNQ1OT1) were ligated into pGPU6/Neo plasmid (GenePharma, Shanghai, China) to silence the expression of KCNQ1OT1, namely sh-KCNQ1OT1. Empty vector and non-targeted shRNAs were used as blank controls. Cell transfection was performed with Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was reversely transcribed into cDNA using Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland). qRT-PCR was performed by using the FastStart Universal SYBR Green Master (Roche, Shanghai, China) according to the manufacturer’s instructions. In addition, total miRNA was extracted using miRNeasy Mini Kit (Qiagen, Shenzhen, China) and reversely transcribed employing TaqMan MicroRNA Reverse Transcription Kit (TaKaRa, Dalian, China). The primers required for qRT-PCR were as follows: KCNQ1OT1: forward, 5ʹ-TTGGTAGGATTTTGTTGAGG-3ʹ and reverse, 5ʹ-CAACCTTCCCCTACTACC-3ʹ; BDNF: forward, 5ʹ-CCACTGCCGGGGATCCGAGA-3ʹ and reverse, 5ʹ-TTTCATGGGCGCCGCCTTCA-3ʹ; GAPDH: forward, 5ʹ-AGGTCGGTGTGAACG-GATTTG-3ʹ and reverse, 5ʹ-GGGGTCGTTGATGGCAACA-3ʹ. GAPDH and U6 were used as the internal references. The data were analyzed with the 2−ΔΔCT method.
MTT Assay
PC-12 cells in the logarithmic growth phase were inoculated in a 96-well plate (1×103 cells/well). The cell viability was measured by the MTT method. When the cells stably grew, different concentrations of ketamine were added, with which the cells were treated for 24 h. Afterward, 10 μL 5mg/mL MTT solution was added into each well, and the cells were incubated for 4 h at 37 °C, and then the MTT-containing medium was removed, followed by the addition of 200 μL dimethyl sulfoxide (DMSO) for dissolving the formazan. Then the plate was shaken for 10 min at 37 °C on a shaker. Next, the optical density (OD) of the cells at 492 nm was measured by a microtiter plate reader (ThermoFisher, Shanghai, China).
TUNEL Assay
To detect the level of apoptosis in the neurons subjected to different treatments, apoptotic cells were labeled with TUNEL and neurons were labeled with NeuN. This procedure was performed in accordance with the instructions of the In Situ Cell Death Detection Kit (Roche, Mannheim, Germany). During the experiment, the slides were protected from light and the nuclei were counterstained with DAPI. The cells were observed and counted under a fluorescence microscope.
Detection of Lactic Dehydrogenase (LDH) Activity
LDH activity was determined using a cytotoxicity detection kit from Roche Applied Science (Indianapolis, IN, USA). OD490nm was measured by spectrophotometer (BioPhotometer; Eppendorf, Hamburg, Germany). LDH in the cells treated with 1% Triton X-100 (Sigma) was used as a positive control with an activity value of 100%.
Dual-Luciferase Reporter Assay
The fragments of KCNQ1OT1, 3ʹUTR of BDNF and their mutant sequences were subcloned into pGL4 vectors (Promega, Madison, WI, USA). Firefly luciferase reporter plasmid containing wild-type or mutant KCNQ1OT1 (pGL4-KCNQ1OT1-WT/MUT) or BDNF 3ʹUTR (pGL4-BDNF-WT/MUT), and renilla luciferase control reporter (Promega, Madison, WI, USA) were co-transfected into PC-12 cells with miR-206 mimics or control miRNAs. After 48 h of culture, the luciferase activity of the cells in each group was measured using the Dual-Luciferase Reporter Assay Kit (Promega, USA).
Enzyme-Linked Immunosorbent Assay (ELISA)
Quantitative analysis of BDNF in samples was performed using a BDNF Emax® ImmunoAssay System (Promega, Madison, WI) according to the manufacturer’s instructions. OD450nm was measured by microplate reader.
Western Blot
Western blot was performed to detect the protein expressions. The cells were lysed with protease inhibitor-containing RIPA lysis buffer (10 μL/mL) (Beyotime, Shanghai, China). Then the mixtures were centrifuged at 14,000×g (4 °C) for 10 min, and the supernatant was collected. Concentration of the protein sample was determined by the BCA protein assay (Beyotime, Shanghai, China). An equal amount of total protein (10 μg/lane) was loaded into sodium dodecyl sulfate polyacrylamide gel and separated by electrophoresis, and then the proteins were transferred onto polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After being blocked with 5% non-fat milk for 1 h at room temperature, the membranes were incubated with the primary antibody anti-BDNF (1:1000, ab108319, Abcam) overnight at 4 °C and then with secondary antibody (Beyotime, Shanghai, China) at room temperature for 1.5 h. Finally, ECL kit (Amersham Pharmacia Biotech, Little Chalfont, UK) was added onto the membranes, and the protein bands in the membranes were visualized using the ChemiDoc XRS imaging system and quantified by Image J image analysis software. The expression of BDNF was normalized to GAPDH on the same membrane.
Data Analysis
All experiments were performed independently for at least three times. Statistical analysis was conducted using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA). Data were expressed as mean ± SD. Two-tailed Student’s t-test was used in this study for making comparison. P < 0.05 was considered to be statistically significant.
Results
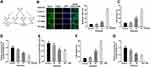
Ketamine Induced the Apoptosis of Neurons and Down-Regulated KCNQ1OT1 Expression
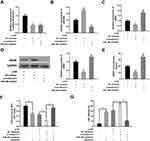
In this study, 7.5, 15 and 30 mg/kg of ketamine was injected into rats, respectively. The neuronal apoptosis was detected by TUNEL staining, and cell viability was tested by LDH kit. The results showed that the number of TUNEL-positive neurons and LDH expression were significantly increased with the presence of high concentrations of ketamine (15 mg/kg and 30 mg/kg), and such effect was more obvious at a higher concentration of ketamine (30 mg/kg) (Figure 1B and C). In addition, results of qRT-PCR showed that KCNQ1OT1 expression was markedly decreased with the increase of ketamine concentration (vs control group) (Figure 1D). To further clarify the effect of ketamine on neurons, we treated PC-12 cells with 25, 50, and 100 μM ketamine in vitro. It was found that the viability of PC-12 cells was observably decreased with the increase of ketamine concentration (Figure 1E), and the LDH release was also significantly increased (Figure 1G). Additionally, the relative expression of KCNQ1OT1 in PC-12 cells was inhibited by ketamine (Figure 1F).
KCNQ1OT1 Overexpression Ameliorated Ketamine-Induced Neuron Injury
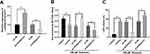
In order to pinpoint the role of KCNQ1OT1 in ketamine-induced neuronal cell injury, we transfected pc-NC, pc-KCNQ1OT1, si-NC and si-KCNQ1OT1 into PC-12 cells to construct KCNQ1OT1 over-expressing and under-expressing cell lines, respectively (Figure 2A). Further experiments revealed that the viability of PC-12 cells with KCNQ1OT1 overexpression was significantly elevated, and LDH expression was markedly decreased under the treatment of ketamine (vs pc-NC group), while the down-regulation of KCNQ1OT1 expression exerted the opposite function (vs si-NC group) (Figure 2B and C).
KCNQ1OT1 Targeted miR-206
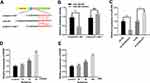
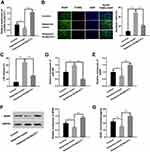
To further fathom the underlying mechanism of KCNQ1OT1 in regulating ketamine-induced neuronal injury, bioinformatics analysis, performed with LncBase Predicted V2.0, was used to predict the binding sites between KCNQ1OT1 and miR-206 (Figure 3A). Dual luciferase reporter assay was performed to further verify the targeting relationship between KCNQ1OT1 and miR-206, the results of which indicated that miR-206 mimics significantly inhibited the luciferase activity of KCNQ1OT1-WT, but failed to reduce that of KCNQ1OT1-MUT (Figure 3B). Further qRT-PCR assay showed that the expression of miR-206 was significantly down-regulated in KCNQ1OT1 overexpressing PC-12 cells, while miR-206 expression was markedly up-regulated in PC-12 cells with KCNQ1OT1 silenced (Figure 3C). These data indicated that miR-206 was a target of KCNQ1OT1 and its expression was negatively regulated by KCNQ1OT1. In addition, qRT-PCR was carried out to detect miR-206 expression under the influence of ketamine. The results showed that, in the hippocampus of the rats treated with 7.5, 15 and 30 mg/kg ketamine and PC-12 cells treated with 25, 50 and 100 μM ketamine, miR-206 expression was observably increased with the increase of ketamine concentration (Figure 3D and E).
Inhibition of miR-206 Attenuated Neuronal Damage Induced by Ketamine
To further verify whether miR-206 participates in ketamine-induced neuronal damage, miR-206 overexpression and down expression cell models were established on PC-12 cell line (Figure 4A). The results showed that inhibition of miR-206 significantly increased the viability of PC-12 cell line and inhibited LDH release while miR-206 overexpression had the opposite effects (Figure 4B and C), revealing that inhibiting miR-206 expression could reduce the damage to neuronal cells caused by ketamine.
MiR-206 Inhibited BDNF Expression in Neurons
Additionally, dual luciferase reporter assay further confirmed that miR-206 could target the 3ʹUTR of BDNF (Figure 5A and B), which was consistent with the previous reports.21,22 Further detection of BDNF expression by qRT-PCR showed that the expression of BDNF in PC-12 cells transfected with miR-206 mimics was significantly lower than in the control group, while in PC-12 cells transfected with miR-206 inhibitors, the expression of BDNF was up-regulated (Figure 5C). In addition, we also detected the mRNA and protein expressions of BDNF in hippocampus of the rats and PC-12 cells treated with different concentrations of ketamine, the findings of which showed that BDNF expression was significantly down-regulated by ketamine, and as the concentration of ketamine increased, this effect became more significant (Figure 5D and E).
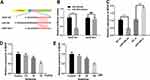
KCNQ1OT1 Attenuated Ketamine-Induced Neuron Injury by Regulating miR-206/BDNF Expression
To further explore the roles of KCNQ1OT1, miR-206, and BDNF in regulating ketamine-induced neuronal injury, we transfected miR-206 inhibitors into PC-12 cells with KCNQ1OT1 knockdown. The results showed that miR-206 inhibitor had no significant effect on KCNQ1OT1 expression, while KCNQ1OT1 down expression elevated miR-206 expression (Figure 6A and B). Furthermore, as shown, the expression levels of BDNF mRNA and protein were inhibited by KCNQ1OT1 knockdown while being significantly up-regulated by miR-206 inhibitors (Figure 6C–E). After the treatment with 100 μM ketamine, the viability of PC-12 cells with KCNQ1OT1 knockdown was significantly decreased, and the release of LDH was markedly increased; however, these phenomena were reversed after the co-transfection of miR-206 inhibitors (Figure 6F and G).
Overexpression of KCNQ1OT1 in Rats Ameliorated Ketamine-Induced Neuronal Injury in vivo
To further verify the protective role of KCNQ1OT1 in ketamine-induced neuronal injury, we established an overexpressing KCNQ1OT1 rat model with adeno-associated virus in vivo (Figure 7A). The results revealed that overexpression of KCNQ1OT1 significantly inhibited the neuronal apoptosis and LDH release (vs control group) (Figure 7B and C). Next, the expressions of KCNQ1OT1, miR-206 and BDNF in brain tissue were further examined. The results showed that overexpression of KCNQ1OT1 attenuated miR-206 expression while upregulating the expressions of BDNF (Figure 7D–G). These results further validated that KCNQ1OT1 could increase BDNF expression by decreasing miR-206 expression, thereby ameliorating the damage to neuronal cells induced by ketamine.
Discussion
Previous studies authenticate that ketamine can induce neuronal apoptosis in developing brain and cultured neurons in vitro,1,8 and even induce the death of human embryonic stem cell-derived neuronal cells.24–28 Moreover, the toxicity of ketamine is positively correlated with dose, and higher doses of ketamine can cause paralysis or even death.29 This study reported for the first time that KCNQ1OT1 could protect against and attenuate ketamine-induced neuronal damage. In terms of the mechanisms, our results indicated that KCNQ1OT1 could down-regulate miR-206 expression and up-regulate BDNF expression.
As a anesthetic, ketamine is widely used clinically, and its influence on neurons has also attracted more and more attention. It is reported that ketamine can be used as an antidepressant at low concentrations.23 Acute administration of ketamine can increase the expression of BDNF that plays an important role in the rapid sustained antidepressant effect of ketamine.30 For instance, injecting a single dose of ketamine (10–15 mg/kg) to rats can increase the protein expression of BDNF in the hippocampus in a short time.31,32 However, a high concentration of ketamine may trigger toxicity to neuronal cells and result in neuronal apoptosis.33,34 Significant neuronal apoptosis is observed 24 h after the treatment with ketamine (100μM), and ketamine can also decrease mitochondrial membrane potential and increase the release of cytochrome c from mitochondria to cytosol.25 In this study, we treated rats and PC-12 cells with different concentrations of ketamine, and the results indicated that the viability of neuronal cells was decreased and the apoptosis was significantly increased with the increment of ketamine concentration, and the release rate of LDH also increased evidently, indicating that neuronal cells were damaged by high concentration of ketamine, which was consistent with the previous reports.
Existing studies unveil that lncRNAs play prominent roles in regulating neuronal damage. For example, lncRNA H19 expression is up-regulated in a hypoxic condition, and miR-28 expression is down-regulated by H19; as a result, SP1, the target gene of miR-28, is up-regulated and ultimately contributes to the deactivation of PDK/AKT and JAK/STAT signaling pathways and protecting nerves from damage caused by hypoxia.35 Additionally, inhibition of lncRNA IGF2AS can protect neural stem cells from neurotoxicity caused by anesthetics via regulating IGF2 expression.36 Here, we found that KCNQ1OT1 expression was significantly down-regulated in ketamine-induced hippocampus tissues and PC-12 cells treated with ketamine. Furthermore, both in vitro and in vivo experiment indicated that overexpressing KCNQ1OT1 significantly reduced ketamine-induced neuronal damage. Thus, KCNQ1OT1 played a neuroprotective role in ketamine-induced neurological damage.
As another class of non-coding RNAs, miRNAs are also found to figure prominently in regulating nerve damage. For example, it is reported that inhibiting miR-124 can reduce ketamine-induced neuronal degeneration.37 It is reported that miRNAs, such as miR-21, miR-34, miR-124, miR-132, and miR-200b, regulate important target genes involved in neuronal apoptosis and neuronal stress-induced adaptation.38 Besides, down-regulation of miR-375 expression protects against ketamine-induced neuronal cell death and neurotoxicity,39 while high miR-206 expression promotes the apoptosis of hippocampal neurons.23 In this study, we found that miR-206 expression was significantly up-regulated by ketamine, and miR-206 overexpression also significantly aggravated neuronal damage. Therefore, we believed that ketamine could aggravate nerve damage by up-regulating miR-206 expression. Additionally, in this study, KCNQ1OT1 was confirmed to target miR-206 to reduce its expression, and this regulatory mechanism partly explained the reason of miR-206 dysregulation during neuronal injury induced by ketamine.
BDNF, a member of the neurotrophic superfamily, plays a prominent role in neuronal maturation, synapse formation, and synaptic plasticity.6 It is reported that the binding of BDNF to tyrosine kinase receptor B (TrkB) can inhibit p75NTR-mediated apoptosis.40 On the other hand, in the developing rat brain, exposure of ketamine can reduce Akt activity and phosphorylation of CREB, resulting in the decreased BDNF expression and neuronal apoptosis.41,42 It is reported that miR-206 is able to regulate nerve regeneration and is involved in the regulation of BDNF expression.23,43,44 Some studies indicate that miR-206 binds to the 3ʹ UTR of BDNF mRNA and thus negatively regulates the expression of BDNF.22,45,46 This study found that BDNF expression in neurons and brain tissue was significantly down-regulated following the treatment of ketamine. Further exploration of the underlying mechanism proved that: 1) BDNF was negatively regulated by up-regulated miR-206 expression following ketamine administration, 2) KCNQIOT1 could significantly increase BDNF expression. Collectively, we supposed that KCNQIOT1 could ameliorate ketamine-induced neuronal injury by inhibiting miR-206 expression and increasing BDNF expression.
Conclusion
Our study shows that KCNQ1OT1 overexpression can ameliorate ketamine-induced nerve damage. The possible mechanism through which KCNQ1OT1 protects neurons is that KCNQ1OT1 can down-regulate miR-206 expression, thus preventing miR-206 from repressing BDNF. This study unveils the key role of KCNQ1OT1/miR-206/BDNF axis in high concentration ketamine-induced neuronal damage, and provides theoretical basis for the clinical application of ketamine.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Statement
All animal procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals from the National Health Research Institutes and Recommendations on Animal Care from the Chinese Neuroscience Society. All of the animal experiments were endorsed by the Institutional Animal Care and Use Committee of Xiangyang Central Hospital (Approval Number: 2017101305; Approval date: October 13, 2017).
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
Co-first authors: Yao Yao, Xuesong Wang.
References
1. Slikker W, Liu F, Rainosek SW, et al. Ketamine-induced toxicity in neurons differentiated from neural stem cells. Mol Neurobiol. 2015;52(2):959–969. doi:10.1007/s12035-015-9248-5
2. Walker SM, Westin BD, Deumens R, Grafe M, Yaksh TL. Effects of intrathecal ketamine in the neonatal rat: evaluation of apoptosis and long-term functional outcome. Anesthesiology. 2010;113(1):147–159. doi:10.1097/ALN.0b013e3181dcd71c
3. Wang C, Liu F, Patterson TA, Paule MG, Slikker W. Relationship between ketamine-induced developmental neurotoxicity and NMDA receptor-mediated calcium influx in neural stem cell-derived neurons. NeuroToxicology. 2017;60:254–259. doi:10.1016/j.neuro.2016.04.015
4. Liu F, Patterson TA, Sadovova N, et al. Ketamine-induced neuronal damage and altered N-methyl-D-aspartate receptor function in rat primary forebrain culture. Toxicol Sci. 2013;131(2):548–557. doi:10.1093/toxsci/kfs296
5. Hotz MA, Del Bino G, Lassota P, Traganos F, Darzynkiewicz Z. Cytostatic and cytotoxic effects of fostriecin on human promyelocytic HL-60 and lymphocytic MOLT-4 leukemic cells. Cancer Res. 1992;52(6):1530.
6. Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci. 2013;14(1):7–23.
7. Zheng X, Lin C, Li Y, Ye J, Zhou J, Guo P. Long noncoding RNA BDNF-AS regulates ketamine-induced neurotoxicity in neural stem cell derived neurons. Biomed Pharmacother. 2016;82:722–728. doi:10.1016/j.biopha.2016.05.050
8. Zuo D, Lin L, Liu Y, et al. Baicalin attenuates ketamine-induced neurotoxicity in the developing rats: involvement of PI3K/Akt and CREB/BDNF/Bcl-2 pathways. Neurotox Res. 2016;30(2):159–172. doi:10.1007/s12640-016-9611-y
9. Smolle MA, Bauernhofer T, Pummer K, Calin GA, Pichler M. Current insights into long non-coding RNAs (LncRNAs) in prostate cancer. Int J Mol Sci. 2017;18(2):473. doi:10.3390/ijms18020473
10. Szybińska A, Leśniak W. P53 dysfunction in neurodegenerative diseases - the cause or effect of pathological changes? Aging Dis. 2017;8(4):506–518. doi:10.14336/AD.2016.1120
11. Chen -L-L, Zhao JC. Functional analysis of long noncoding RNAs in development and disease. In: Yeo GW, editor. Systems Biology of RNA Binding Proteins. New York: Springer New York; 2014:129–158.
12. Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69–80. doi:10.1016/j.brainresbull.2013.06.001
13. Charannya Sozheesvari S, Qidong H. Non-coding RNA in brain development and disorder. Curr Med Chem. 2017;24(18):1983–1997.
14. Zhong J, Jiang L, Huang Z, et al. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav Immun. 2017;65:183–194. doi:10.1016/j.bbi.2017.05.001
15. Liang Z, Chi YJ, Lin GQ, Xiao LF, Su GL, Yang LM. LncRNA MEG3 participates in neuronal cell injury induced by subarachnoid hemorrhage via inhibiting the Pi3k/Akt pathway. Eur Rev Med Pharmacol Sci. 2018;22(9):2824–2831.
16. Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi:10.1016/j.molcel.2008.08.022
17. Li X, Dai Y, Yan S, et al. Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute myocardial infarction. Biochem Biophys Res Commun. 2017;491(4):1026–1033. doi:10.1016/j.bbrc.2017.08.005
18. Zhou S, Li M, Zeng D, et al. A single nucleotide polymorphism in 3ʹ untranslated region of epithelial growth factor receptor confers risk for pulmonary hypertension in chronic obstructive pulmonary disease. Cell Physiol Biochem. 2015;36(1):166–178. doi:10.1159/000374061
19. Carrillo ED, Sampieri R, Hernández A, García MC, Sánchez JA. MiR-132 regulates rem expression in cardiomyocytes during long-term β-adrenoceptor agonism. Cell Physiol Biochem. 2015;36(1):141–154. doi:10.1159/000374059
20. Cao SE, Tian J, Chen S, Zhang X, Zhang Y. Role of miR-34c in ketamine-induced neurotoxicity in neonatal mice hippocampus. Cell Biol Int. 2015;39(2):164–168. doi:10.1002/cbin.10349
21. Solomon MG, Griffin WC, Lopez MF, Becker HC. Brain regional and temporal changes in BDNF mRNA and microRNA-206 expression in mice exposed to repeated cycles of chronic intermittent ethanol and forced swim stress. Neuroscience. 2019;406:617–625. doi:10.1016/j.neuroscience.2019.02.012
22. Tapocik JD, Barbier E, Flanigan M, et al. microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J Neurosci. 2014;34(13):4581–4588. doi:10.1523/JNEUROSCI.0445-14.2014
23. Yang X, Yang Q, Wang X, et al. MicroRNA expression profile and functional analysis reveal that miR-206 is a critical novel gene for the expression of BDNF induced by ketamine. Neuromolecular Med. 2014;16(3):594–605. doi:10.1007/s12017-014-8312-z
24. Ito H, Uchida T, Makita K, Rodrigo F, Franco R. Ketamine causes mitochondrial dysfunction in human induced pluripotent stem cell-derived neurons. PLoS One. 2015;10(5):e0128445. doi:10.1371/journal.pone.0128445
25. Bai X, Yan Y, Canfield S, et al. Ketamine enhances human neural stem cell proliferation and induces neuronal apoptosis via reactive oxygen species–mediated mitochondrial pathway. Anesth Analg. 2013;116(4):869–880. doi:10.1213/ANE.0b013e3182860fc9
26. Bai X, Twaroski D, Bosnjak ZJ. Modeling anesthetic developmental neurotoxicity using human stem cells. Semin Cardiothorac Vasc Anesth. 2013;17(4):276–287. doi:10.1177/1089253213495923
27. Ye Z, Li Q, Guo Q, et al. Ketamine induces hippocampal apoptosis through a mechanism associated with the caspase-1 dependent pyroptosis. Neuropharmacology. 2018;128:63–75. doi:10.1016/j.neuropharm.2017.09.035
28. Wang Q, Shen FY, Zou R, Zheng JJ, Yu X, Wang YW. Ketamine-induced apoptosis in the mouse cerebral cortex follows similar characteristic of physiological apoptosis and can be regulated by neuronal activity. Mol Brain. 2017;10(1):24. doi:10.1186/s13041-017-0302-2
29. Muetzelfeldt L, Kamboj SK, Rees H, Taylor J, Morgan CJA, Curran HV. Journey through the K-hole: phenomenological aspects of ketamine use. Drug Alcohol Depend. 2008;95(3):219–229. doi:10.1016/j.drugalcdep.2008.01.024
30. Frey BN, Andreazza AC, Ceresér KM, et al. Effects of mood stabilizers on hippocampus BDNF levels in an animal model of mania. Life Sci. 2006;79(3):281–286. doi:10.1016/j.lfs.2006.01.002
31. Garcia LS, Comim CM, Valvassori SS, et al. Acute administration of ketamine induces antidepressant-like effects in the forced swimming test and increases BDNF levels in the rat hippocampus. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(1):140–144. doi:10.1016/j.pnpbp.2007.07.027
32. Garcia LS, Comim CM, Valvassori SS, et al. Chronic administration of ketamine elicits antidepressant-like effects in rats without affecting hippocampal brain-derived neurotrophic factor protein levels. Basic Clin Pharmacol Toxicol. 2008;103(6):502–506. doi:10.1111/j.1742-7843.2008.00210.x
33. Gutierrez S, Carnes A, Finucane B, et al. Is age-dependent, ketamine-induced apoptosis in the rat somatosensory cortex influenced by temperature? Neuroscience. 2010;168(1):253–262. doi:10.1016/j.neuroscience.2010.03.016
34. Turner CP, Gutierrez S, Liu C, et al. Strategies to defeat ketamine-induced neonatal brain injury. Neuroscience. 2012;210:384–392. doi:10.1016/j.neuroscience.2012.02.015
35. Chen Z, Chen X, Guo R, Meng J. Protective effects of lncRNA H19 silence against hypoxia-induced injury in PC-12 cells by regulating miR-28. Int J Biol Macromol. 2019;121:546–555. doi:10.1016/j.ijbiomac.2018.10.033
36. Song C, Song C, Chen K, Zhang X. Inhibition of long non-coding RNA IGF2AS protects apoptosis and neuronal loss in anesthetic-damaged mouse neural stem cell derived neurons. Biomed Pharmacother. 2017;85:218–224.
37. Xu H, Zhang J, Zhou W, Feng Y, Teng S, Song X. The role of miR-124 in modulating hippocampal neurotoxicity induced by ketamine anesthesia. Int J Neurosci. 2015;125(3):213–220. doi:10.3109/00207454.2014.919915
38. Sessa F, Maglietta F, Bertozzi G, et al. Human brain injury and miRNAs: an experimental study. Int J Mol Sci. 2019;20(7):1546. doi:10.3390/ijms20071546
39. Zhao X, Shu F, Wang X, et al. Inhibition of microRNA-375 ameliorated ketamine-induced neurotoxicity in human embryonic stem cell derived neurons. Eur J Pharmacol. 2019;844:56–64. doi:10.1016/j.ejphar.2018.11.035
40. Davey F, Davies AM. TrkB signalling inhibits p75-mediated apoptosis induced by nerve growth factor in embryonic proprioceptive neurons. Curr Biol. 1998;8(16):915–918. doi:10.1016/S0960-9822(07)00371-5
41. Zuo D, Sun F, Cui J, et al. Alcohol amplifies ketamine-induced apoptosis in primary cultured cortical neurons and PC12 cells through down-regulating CREB-related signaling pathways. Sci Rep. 2017;7(1):10523. doi:10.1038/s41598-017-10868-z
42. Lu LX, Yon JH, Carter LB, Jevtovic-Todorovic V. General anesthesia activates BDNF-dependent neuroapoptosis in the developing rat brain. Apoptosis. 2006;11(9):1603–1615. doi:10.1007/s10495-006-8762-3
43. Williams AH, Valdez G, Moresi V, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science (New York, NY). 2009;326(5959):1549–1554. doi:10.1126/science.1181046
44. Miura P, Amirouche A, Clow C, Belanger G, Jasmin BJ. Brain-derived neurotrophic factor expression is repressed during myogenic differentiation by miR-206. J Neurochem. 2012;120(2):230–238. doi:10.1111/j.1471-4159.2011.07583.x
45. Lee ST, Chu K, Jung KH, et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72(2):269–277. doi:10.1002/ana.23588
46. Tapocik JD, Solomon M, Flanigan M, et al. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenomics J. 2013;13(3):286–296. doi:10.1038/tpj.2012.17
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.