Back to Journals » Cancer Management and Research » Volume 12
LncRNA HCP5 Stimulates the Proliferation of Non-Small Cell Lung Cancer Cells by Up-Regulating Survivin Through the Down-Regulation of miR-320
Authors Li C, Lei Z, Peng B, Zhu J, Chen L
Received 6 July 2019
Accepted for publication 10 October 2019
Published 14 February 2020 Volume 2020:12 Pages 1129—1134
DOI https://doi.org/10.2147/CMAR.S222221
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lu-Zhe Sun
Chao Li,1 Zhang Lei,2 Bin Peng,1 Jiang Zhu,1 Li Chen3
1Oncology Department, Second People’s Hospital of Jingmen, Jingmen City, Hubei Province 448000, People’s Republic of China; 2Department of Oncology, The Central Hospital of Wuhan Affiliated to Tongji Medical College, Huazhong University of Science and Technology, Wuhan City, Hubei Province 430061, People’s Republic of China; 3Department of Traditional Chinese Medicine, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Art and Science, Xiangyang City, Hubei Province 441021, People’s Republic of China
Correspondence: Li Chen
Department of Traditional Chinese Medicine, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Art and Science, Jingzhou Street 136#, Xiangyang City, Hubei Province 441021, People’s Republic of China
Tel +86 0710-3522791
Email [email protected]
Introduction: We explored the roles of lncRNA HCP5 in non-small cell lung cancer (NSCLC).
Methods: Levels of HCP5 were measured by performing qPCR and data were compared between non-tumor and NSCLC tissue samples by performing a paired t-test. Expression levels of miR-320 and survivin mRNA in NSCLC tissues were also measured by performing qPCR. The effects of HCP5, miR-320 and survivin overexpression on the proliferation of H23 cells were analyzed by cell proliferation assay.
Results: We found that HCP5 was up-regulated in NSCLC and predicted the poor survival of NSCLC patients. HCP5 was negatively correlated with miR-320 but positively correlated with survivin in NSCLC tissues. In NSCLC cells, HCP5 overexpression led to the up-regulated survivin and down-regulated miR-320. Moreover, miR-320 overexpression failed to affect HCP5 but down-regulated survivin. Cell proliferation assay showed that HCP5 and survivin overexpression led to increased, while miR-320 overexpression led to decreased cell proliferation rate. In addition, miR-320 overexpression reduced the effects of HCP5 overexpression.
Conclusion: Therefore, HCP5 may stimulate the proliferation of NSCLC cells by up-regulating survivin through the down-regulation of miR-320.
Keywords: HCP5, non-small cell lung cancer, miR-320, survivin
Introduction
Lung cancer has been the most commonly diagnosed malignancy for decades.1 In 2018, 2,093,876 new lung cancer cases were diagnosed, which account for 11.6% of all cancers.2 During the same period, 1,761,007 people died of lung cancer, which accounts for about 18.4% of all cancer deaths.2 About 85% of lung cancer patients are non-small cell lung cancer (NSCLC), which is further divided into adenocarcinoma and squamous cell carcinoma two subtypes.1 Smoking is the main risk factor of NSCLC.3 However, NSCLC also affects never-smokers.4 In addition, NSCLC is usually diagnosed at advanced stages and prognosis is generally poor.5 Therefore, more effective therapeutic approaches are awaited to improve the survival of NSCLC patients.
A wealth of evidence has shown that genetic alterations are critical players in the pathogenesis of NSCLC.6,7 Survivin belongs to the family of the inhibitor of apoptosis. The major function of survivin is to inactivate caspases and suppress cell death, thereby promoting tumor growth and metastasis.8 Therefore, inactivation of survivin is considered as a potential therapeutic target for cancer therapies.9 It has been reported that miR-320 can target survivin to participate in the function of insulin in myocardial ischemia.10 However, the interaction between miR-320 and survivin in cancer biology is unclear. Long (>200nt) non-coding RNA (lncRNA) HCP5 has been characterized as an oncogenic lncRNA in follicular thyroid carcinoma and cervical cancer.11,12 Our preliminary bioinformatics analysis showed that HCP5 can form base pairing with the precursor of miR-320, which indicates the potential interaction between these two. This study aimed to investigate the interactions among HCP5, miR-320, and survivin in NSCLC.
Materials and Methods
NSCLC Patients
This study passed the review of Xiangyang Central Hospital Ethics Committee. A total of 166 patients with NSCLC were admitted by Xiangyang Central Hospital between March 2011 and April 2014. From those patients, 63 patients (44 males and 19 females; 38 to 70 years; 53.1 ± 7.3 years) were enrolled in this study. Inclusion criteria: 1) adenocarcinoma cases; 2) newly diagnosed cases; 3) no therapies initiated. Exclusion criteria: 1) other types of NSCLC; 2) recurrent NSCLC; 3) other clinical disorders were observed. All the 63 patients were informed of experimental details and informed consent was signed by all patients.
Clinical Staging and Specimen Collections
All the 63 patients were staged by clinical findings and AJCC staging system. The results revealed 29 and 34 cases at stage III and IV.
All patients were diagnosed by lung histopathological biopsy. The biopsy was performed under the guidance of MRI to collect both non-tumor lung tissue and NSCLC tissues. All tissue specimens were confirmed by histopathological exams.
Follow-Up Study
From the day of admission, all patients were followed up for 5 years or until their death. Patients were visited monthly through telephone and/or outpatient visits. Survival conditions were recorded and used in survival analysis.
NSCLC Cell Line and Cell Transfections
H23 human NSCLC (adenocarcinoma) cell line (ATCC, USA) was used as an NSCLC cell model. A mixture of 10% FBS and 90% RPMI-1640 medium was used as a cell culture medium. Cell culture conditions were 37 °C, 95 humidity and 5% CO2.
Vectors expressing HCP5 and survivin were constructed using pcDNA3 vector, and the vector constructions were completed by Sangon (Shanghai, China). Negative control (NC) miRNA and miR-320 mimic were synthesized by Sangon. H23 cells were harvested at the confluence of 75% and Lipofectamine 2000 (Sangon) was used to transfect 10 nM vectors (empty vector as NC group) or 50 nM miRNAs (NC miRNA as NC group) into 106 cells. In all transfections, untransfected cells were control (C) cells. The following experiments were performed using cells harvested at 48h post-transfection.
Total RNA Extractions
H23 cells were harvested at 48h post-transfection and cells were counted. Tissue samples (0.04g per sample) were ground in liquid nitrogen to make a fine powder. Total RNAs in cells and tissues were extracted using Ribozol (Sigma-Aldrich). Eighty-five percent ethanol was used to precipitate and wash RNA samples to harvest miRNAs.
qPCR
Expression levels of HCP5 and survivin were measured by performing qPCR. Transcriptions were performed using TruScript Reverse Transcriptase Kit (Norgenbiotek) and KAPA SYBR FAST qPCR Kit (Roche) were used to prepare qPCR mixtures using 18S rRNA as the endogenous control.
In order to measure the expression levels of miR-320, All-in-One™ miRNA qRT-PCR Reagent Kit (GeneCopoeia) was used to perform poly(A) addition, reverse transcriptions and qPCR assays.
Primer sequences were: 5ʹ-CCACTATTGGCCATCAAAGG-3ʹ (Forward) and 5ʹ-ATACTGTCCAATTCCCCTGT-3ʹ (reverse) for human HCP5; 5ʹ-CTACCACATCCAAGGAAGC-3ʹ (Forward) and 5ʹ-TTTTCGTCACTACCTCCCCG-3ʹ (reverse) for human 18S rRNA. The forward primer of miR-320 was: 5ʹ-GCCTTCTCTTCCCGGTTCT-3ʹ. Reverse primer of miR-320 and U6 primers were included in the kit.
The 2−ΔΔCT method was used to process all Ct values. All PCR reactions were repeated 3 times.
Western Blot
The effects of HCP5 and miR-320 overexpression on the expression of survivin were analyzed by Western blot. H23 cells were harvested at 48h post-transfection and cells were counted. Total proteins were extracted using RIPA solution (Sangon). To denature proteins, all protein samples were incubated in boiling water for 10 min. Twelve percent SDS-PAGE gel was used to perform electrophoresis, followed by gel transfer to PVDF membranes. Membranes were first incubated with primary antibodies of rabbit anti-survivin (1:2000, ab469, Abcam) and anti-GAPDH (1:1500, ab37168, Abcam), and the incubation was performed at 4°C for 18h. The secondary antibody was goat HRP (IgG) (1:1800; ab6721; Abcam) and the incubation was performed at 24°C for 2h. ECL solution (Sigma-Aldrich) was used to develop signals. Image J v1.46 software was used to normalize signals.
Cell Proliferation Analysis
The effects of transfections on the proliferation of H23 cells were analyzed by cell proliferation assay. Briefly, 3×104 cells were mixed with 1 mL mixture of 90% RPMI-1640 medium and 10% FBS to prepare single-cell suspensions, followed by cell culture in cell plate (96-well, 0.1mL per well) under aforementioned conditions. Each well was added with 10ul CCK-8 solution (Sigma-Aldrich) at 4h before the end of cell culture. OD values were measured at 450 nm.
Statistical Analysis
Three biological replicates were included in each experiment. All data analyses were performed using the mean values of the replicates. Correlations were analyzed by performing Pearson’s correlation coefficient. Differences were explored between two types of samples (NSCLC vs non-tumor) and among cell transfection groups. Differences among multiple groups were explored by performing ANOVA (one-way) and Tukey’s test. To perform survival analysis, the 63 patients were grouped into high (n=32) and low (n=31) HCP5 level groups with the median expression level of HCP5 in NSCLC tissues as the cutoff value. K-M plotter and log-rank test were used to plot and compare survival curves. p<0.05 was statistically significant.
Results
HCP5 Was Up-Regulated in NSCLC and Predicted the Survival of NSCLC Patients
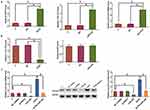
Levels of HCP5 were measured by performing qPCR and data were compared between non-tumor and NSCLC tissue samples by performing a paired t-test. Comparing to non-tumor tissues, expression levels of HCP5 were significantly higher in NSCLC (Figure 1A, p<0.05). Survival curves were plotted and compared using aforementioned methods. Comparing to patients in low HCP5 level group, the overall survival rate of patients in high HCP5 level group was significantly lower (Figure 1B).
HCP5 Was Negatively Correlated with miR-320 but Positively Correlated with Survivin in NSCLC Tissues
Expression levels of miR-320 and survivin mRNA in NSCLC tissues were also measured by performing qPCR. Correlations between HCP5 and miR-320/survivin were analyzed by performing Pearson’s correlation coefficient. It was observed that HCP5 was negatively correlated with miR-320 (Figure 2A) but positively correlated with survivin in NSCLC tissues (Figure 2B).
HCP5 Up-Regulate Survivin Through miR-320
To analyze the interactions among HCP5, miR-320, and survivin, H23 cells were transfected with HCP5 expression vector, miR-320 mimic and survivin expression vector, and the overexpression of HCP5, miR-320 and survivin were confirmed by qPCR at 48h post-transfection. Comparing to C and NC (NC miRNA or empty pcDNA3 vector) groups, expression levels of HCP5, miR-320 and survivin mRNA were significantly increased at 48h post-transfection (Figure 3A, p<0.05). Comparing to two controls, HCP5 overexpression led to down-regulated miR-320, while miR-320 overexpression failed to affect HCP5 (Figure 3B, p<0.05). In addition, HCP5 overexpression led to up-regulated survivin. Moreover, miR-320 overexpression played the opposite role and attenuated the effects of HCP5 overexpression (Figure 3C, p<0.05).
HCP5 Promoted the Proliferation of H23 Cells Through miR-320 and Survivin
The effects of HCP5, miR-320 and survivin overexpression on the proliferation of H23 cells were analyzed by cell proliferation assay. Comparing to two controls, cell proliferation assay showed that HCP5 and survivin overexpression led to increased, while miR-320 overexpression led to decreased cell proliferation rate. In addition, miR-320 overexpression reduced the effects of HCP5 overexpression (Figure 4, p<0.05).
Discussion
The expression pattern and functions of HCP5 in NSCLC have been investigated in the present study. We found that HCP5 was up-regulated in NSCLC and predicted poor survival. In addition, HCP5 may up-regulate survivin by down-regulating miR-320 to promote the proliferation of NSCLC cells.
The functions of HCP5 have been investigated in follicular thyroid carcinoma and cervical cancer.11,12 HCP5 was up-regulated in both types of cancers and promote cancer development either by sponging tumor-suppressive miRNAs or down-regulation tumor-suppressive miRNAs.11,12 In this study, we first reported the up-regulation of HCP5 in NSCLC and the increased cell proliferation rate of NSCLC cells after HCP5 overexpression. Our data revealed the oncogenic functions of HCP5 in NSCLC.
As we just mentioned, HCP5 participates in cancer biology mainly by interacting with miRNAs.11,12 In this study, we proved that HCP5 can down-regulate miR-320 expression. However, the specific mechanism is unclear. It has been well established that the hairpin structure of the miRNA precursor is critical for the maturation of miRNAs.13–15 Our preliminary bioinformatics analysis showed that HCP5 may form base pairing with the miR-320 precursor (Supplementary Figure 1). Therefore, HCP5 may disturb the maturation of miR-320 to down-regulate the level of mature miR-320. MiR-320 can target survivin to regulate the functions of insulin in myocardial ischemia.10 In the present study, we also observed the down-regulation of survivin in NSCLC cells after miR-320 overexpression. Therefore, miR-320 may also target survivin in NSCLC cells. In addition, we characterized a novel HCP5/miR-320/survivin pathway in NSCLC.
Accurate prognostic assignment plays a critical role in the prognosis of NSCLC.16,17 In this study, we showed that high levels of HCP5 expression were closely correlated with the poor survival of NSCLC patients. However, the clinical application of HCP5 as a prognostic factor for NSCLC remains to be further tested by more clinical trials.
It is worth noting that HCP5 was reported to be down-regulated in lung adenocarcinoma, a subtype of NSCLC.18 However, by analyzing the TCGA dataset we observed the up-regulation of HCP5 in lung adenocarcinoma (25.19 vs 21.47). Our data are consistent with the TCGA dataset. However, more studies are needed to further confirm the conclusions. Survivin inhibits the growth of tumors by inhibiting cell apoptosis.8,9 Our study confirmed the enhancing effects of survivin on NSCLC proliferation. Based on the data presented in this study, regulation of HCP5 may indirectly regulate the expression of survivin to affect NSCLC development and progression.
In conclusion, HCP5 is up-regulated in NSCLC and it may promote the proliferation of NSCLC cells by up-regulating survivin through the down-regulation of miR-320.
Acknowledgments
We acknowledge financial support from Jingmen Science and Technology Project YFZD2017044, Scientific Research Project of Hubei Provincial Health and Health Commission WJ2015MB206 the Natural Science Foundation of Hubei Province China under Grant 2017CFB408.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of Xiangyang Central Hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients and healthy volunteers provided written informed consent prior to their inclusion within the study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454.
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
3. Bernatsky S, Ramsey-Goldman R, Petri M, et al. Smoking is the most significant modifiable lung cancer risk factor in systemic lupus erythematosus. J Rheumatol. 2018;45(3):393–396. doi:10.3899/jrheum.170652
4. Gazdar AF, Zhou C. Lung cancer in never-smokers: a different disease[M]/IASLC Thoracic Oncology. Content Repository Only. 2018;23–29:e3.
5. Fernandez FG, Kosinski AS, Furnary AP, et al. Differential effects of operative complications on survival after surgery for primary lung cancer. J Thorac Cardiovasc Surg. 2018;155(3):1254–1264. e1. doi:10.1016/j.jtcvs.2017.09.149
6. Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54(1):73–87. doi:10.1146/annurev.med.54.101601.152202
7. Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123(1):1–7. doi:10.1002/(ISSN)1097-0215
8. Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8(1):61–70. doi:10.1038/nrc2293
9. Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3(1):46–54. doi:10.1038/nrc968
10. Yang N, Wu L, Zhao Y, et al. MicroRNA‐320 involves in the cardioprotective effect of insulin against myocardial ischemia by targeting survivin. Cell Biochem Funct. 2018;36(3):166–171. doi:10.1002/cbf.v36.3
11. Liang L, Xu J, Wang M, et al. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018;9(3):372. doi:10.1038/s41419-018-0382-7
12. Yu Y, Shen HM, Fang DM, et al. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur Rev Med Pharmacol Sci. 2018;22(15):4812–4819. doi:10.26355/eurrev_201808_15616
13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
14. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi:10.1038/nrm3838
15. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi:10.1038/nrm1644
16. Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi:10.1016/j.ccr.2006.01.025
17. Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non–small-cell lung cancer. N Engl J Med. 2006;355(6):570–580. doi:10.1056/NEJMoa060467
18. Zhu TG, Xiao X, Wei Q, et al. Revealing potential long non-coding RNA biomarkers in lung adenocarcinoma using long non-coding RNA-mediated competitive endogenous RNA network. Braz J Med Biol Res. 2017;50(9):e6297. doi:10.1590/1414-431X20176071
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.